Abstract
MADS-domain transcription factors have been shown to act as key repressors or activators of the transition to flowering and as master regulators of reproductive organ identities. Despite their important roles in plant development, the origin of several MADS-box subfamilies has remained enigmatic so far. Here we demonstrate, through a combination of genome synteny and phylogenetic reconstructions, the origin of three major, apparently angiosperm-specific MADS-box gene clades: FLOWERING LOCUS C- (FLC-), SQUAMOSA- (SQUA-) and SEPALLATA- (SEP-) -like genes. We find that these lineages derive from a single ancestral tandem duplication in a common ancestor of extant seed plants. Contrary to common belief, we show that FLC-like genes are present in cereals where they can also act as floral repressors responsive to prolonged cold or vernalization. This opens a new perspective on the translation of findings from Arabidopsis to cereal crops, in which vernalization was originally described.
Similar content being viewed by others
Introduction
Flowering plants have evolved an enormous complexity and diversity in the developmental transition from vegetative to reproductive growth. Environmental and internal cues are integrated in a quantitative flowering response that varies between species and even between ecotypes1. Plants growing in temperate climates use photoperiod or day-length, in addition to vernalization or low temperatures to sense the passing of winter into optimal reproductive environmental conditions2. The elaboration of reproductive development in flowering plants is associated with the origin and diversification of developmental control genes, most prominently members of the MADS-box transcription factor family. The origin of several subfamilies of MADS-box genes with crucial roles in the floral transition remains shrouded in mystery, in that they appear to be present in just flowering plants or in specific lineages of flowering plants.
One lineage of MADS-box genes with a highly enigmatic origin is the clade of FLOWERING LOCUS C (FLC) genes. In the model plant Arabidopsis thaliana, FLC is a central repressor of the floral transition3, where it inhibits flowering by directly repressing the activity of central flowering promoters, namely SUPPRESSOR OF OVEREXPRESSION OF CONSTANS (SOC1), FLOWERING LOCUS D (FD) and FLOWERING LOCUS T (FT)1,2,3,4,5. Vernalization alleviates this repression by negatively regulating FLC expression through epigenetic modifications of the chromatin structure at the FLC locus2,4. FLC has five closely related paralogs in the Arabidopsis thaliana genome, some of which also act as floral repressors6,7,8. These paralogs arose in evolution through sequential tandem and genome duplications within the order Brassicales9,10. Tandem duplications of FLC-like genes are not uncommon, as they have also been reported in other Arabidopsis species10. Outside of Brassicales FLC-like genes have been identified in more distantly related core eudicot species. For instance in sugar beet (Beta vulgaris), a crop with a strong vernalization requirement, FLC expression also responds to vernalization11. FLC homologues, however, have not been identified outside the core eudicots and the phylogenetic position of this subfamily in the larger MADS-box gene phylogeny, and therefore its evolutionary origin, is uncertain. Historically, vernalization has been extensively studied in temperate monocot crops, like winter varieties of wheat and barley12. To significantly accelerate flowering and subsequent seed set, winter cereals require a sufficiently long period of cold. In contrast to Arabidopsis, however, their vernalization response involves other members of the MADS-box gene family, as well as other genes12,13,14. Therefore, it has previously been suggested that the vernalization response in temperate cereals and eudicots may have evolved independently13,14,15.
FLC-like genes are not the only subfamily of MADS-box genes with an enigmatic origin. While members of the SQUAMOSA (SQUA) and SEPALLATA (SEP) subfamilies have been identified in all major flowering plant lineages, no gymnosperm representatives have so far been found despite the availability of extensive transcriptome investigations and targeted cloning efforts16,17. In angiosperms, rounds of polyploidization (whole-genome duplications) probably generated many of the observed gene duplications in the SQUA and SEP subfamilies18,19,20,21. Members of the SQUA subfamily are generally positive regulators of the floral transition as they control the formation of inflorescence and floral meristems22. SEP genes act as key regulators of floral organ specification, and in a partially redundant manner with SQUA-like genes in floral meristem specification23,24.
Clarifying the origin of SEP, SQUA and FLC subfamilies can greatly contribute to our understanding of the evolution of flowering plants. In this study, we combined genomic synteny-based approaches and phylogeny reconstruction to understand the evolutionary history of these MADS-box gene subfamilies. This allowed us to identify FLC orthologs in monocots. Similar to Arabidopsis FLC, the expression of Brachypodium FLC-like genes is responsive to a prolonged cold period. The tandem arrangement of the FLC, SEP and SQUA subfamilies suggests an origin of these subfamilies by an ancient tandem duplication before the origin of extant flowering plants, followed by segmental duplications linked to rounds of polyploidization. Our results close an important gap in our understanding on the origin of developmental key regulatory genes in flowering plants.
Results
Conserved SEP3-FLC and SEP1-SQUA tandems in core eudicots
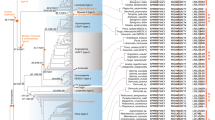
The conservation of gene order between species and between duplicated genomic segments can provide insights into the evolutionary history of genes. Such information can complement phylogenies as an independent source of evidence for evolutionary relationships between paralogous (duplicated) gene lineages25, including MADS-box genes26. Therefore, we studied genomic locations of MIKC-type MADS-box genes in phylogenetically informative flowering plant genomes. To this end, we identified evolutionary conserved tandem arrangements for members of different MADS-box gene subfamilies. We observed that members of the SEP1 and SQUA, as well as SEP3 and FLC subfamilies are arranged in tandem in several core eudicot genomes (Fig. 1; Supplementary Table S1). While members of the SQUA subfamily are exclusively next to members of the SEP1 subclade, FLC genes are consistently next to members of the SEP3 subclade.
Genes that exhibit conserved synteny throughout the angiosperms are represented by coloured arrows and are connected by vertical lines that are defined at the top of the figure. Loci in white did not exhibit synteny and are not discussed. SBP-like genes belonging to different monophyletic groups are differentiated by a different grey tone. On the left side of each genome fragment, position coordinates are shown.
It has been suggested that core eudicots evolved from an ancient hexaploid ancestor27. As such, numerous MADS-box subfamilies exhibit three core eudicot-specific subclades, for example in the SQUA and SEP1 clades19,21. In agreement with this, we observe that in Vitis, Populus and Solanum the SEP1-SQUA tandem is also present in triplicate corresponding to the different core eudicot-specific subclades (Supplementary Table S1).
To understand whether the genomic arrangements of MADS-box genes from the different subfamilies reflect a common evolutionary origin, we performed phylogenetic analyses including a representative sampling of subfamily members (Fig. 2). We found that FLC-like genes are sister to SQUA genes in a monophyletic group that receives strong support (95 maximum-likelihood bootstrap support (BS), 1.00 Bayesian posterior probability (BPP)). Similarly, the SEP3 and SEP1 subfamilies form a monophyletic group (99 BS, 0.97 BPP), as had been observed previously28. SEP-like genes together with gymnosperm and angiosperm AGL6-like genes form a supported monophyletic group (83 BS, 0.96 BBP). These gene lineages are all nested within a strongly supported superfamily, which includes SEP-, AGL6-, SQUA- and FLC-like genes (93 BS, 0.97 BPP). Phylogenetic analysis therefore supports the idea of a common origin of these subfamilies by combination of tandem and segmental duplications.
FLOWERING LOCUS C orthologs are present in monocots
SQUA and SEP1 tandem arrangements can also be identified in monocot genomes (Fig. 1.). Interestingly, in monocots we identified tandems of SEP3 with MADS-box genes that are currently annotated as ‘monocot-specific’ and being related to type I or MIKC*-type MADS-box genes29,30,31, which are evolutionarily more ancient and structurally different lineages of MADS-box genes. The genomic position of these genes, however, suggests that they represent members of the FLC subfamily in monocots. Indeed, in the phylogeny these enigmatic monocot genes group with FLC genes from core eudicots with high BS (99 BS, 1.00 BPP, Fig. 2). This indicates that these genes were previously misclassified FLC orthologs in monocots. The FLC monocot clade itself is strongly supported (100 BS, 1.00 BPP) and consists solely of genes from Poaceae, except for MpFLC (Musa paradisiaca, Musaceae). In the order Poales, we identified two major FLC clades, which we will refer to as OsMADS37-like genes and OsMADS51-like genes. The fact that both rice and sorghum have duplicate FLC copies could be explained by the putative whole-genome duplication 56–72 million years ago (mya) termed ‘rho’, which occurred before the divergence of the major grass lineages25,32. Subsequently, in one of these gene lineages a tandem duplication occurred, apparently before the origin of Pooideae. These monocot-specific duplications resulted in three FLC clades in temperate grasses and two major lineages in the order Poales33. The monocot FLC genes are characterized by divergent, short protein sequences, which probably made it difficult to identify them through traditional similarity searches, such as Basic Local Alignment Searches Tool (BLAST)11.
In recent studies, several members of the OsMADS51 clade have been shown to function in vernalization-controlled flowering in temperate grasses similar to FLC-like genes in core-eudicots (Supplementary Fig. S1) (refs 33, 34). To our knowledge, no members of the OsMADS37 clade have previously been investigated. To understand the cold regulation of the different FLC paralogs in more detail, we monitored their expression level in the temperate grass model Brachypodium distachyon after 2, 4 and 6 weeks of prolonged cold (4 °C) using quantitative reverse transcription-PCR (qRT–PCR). The expression of BdMADS37 (Bradi3g41297) decreases gradually during vernalization (Fig. 3a). The expression of the OsMADS51-like gene BdODDSOC2 (Bradi2g59190) is more rapidly downregulated by vernalization and a minimal expression level is reached already after 2 weeks of cold, or possibly sooner, and remained stable in subsequent weeks (Fig. 3c). In contrast, the expression of BdODDSOC1 (Bradi2g59120) at 4 °C mirrored the control condition during 2 and 4 weeks cold exposure, but increased after 6 weeks in vernalized plants compared with control plants (Fig. 3b). In conclusion, Brachypodium FLC-like paralogs are responsive to prolonged cold exposure, but the nature of the response differs both qualitatively and quantitatively between paralogs.
Expression of FLC-like genes in Brachypodium were monitored using qRT–PCR in plants exposed to a 6-week-period at 4 °C (vernalized) or 28 °C (control). Expression in vernalized samples is represented by diamonds, while the control samples are indicated by squares. Prevern, prevernalization; 2w, two weeks; 4 w, four weeks; 6 w, six weeks. Error bars represent the s.e.m. of three biological replicates.
Ancient duplication events in the MADS-box gene superclade
The phylogenetic sister-relationship between the FLC/SQUA and SEP3/SEP1 clades, in combination with the SEP1-SQUA and SEP3-FLC tandem arrangements suggest a single origin of these subfamilies through ancient tandem duplication. The presence of gymnosperm AGL6-like genes in this large superfamily of flowering time genes indicates that this tandem duplication predates the divergence of gymnosperms and angiosperms. The tandem arrangements can provide a simple explanation for the apparent absence of SEP-, SQUA- and FLC-like genes in extant gymnosperms after the split of AGL6 and SEP clades, through a single segmental deletion of the ancestral SEP-SQUA/FLC tandem along the gymnosperm stem-lineage. In agreement with the ancient evolutionary origin of the tandem arrangements, we observed that the SEP1-SQUA tandem is also present in the Amborella genome, the species sister to all other flowering plants (Fig. 1). However, in Amborella neither FLC-like MADS-box genes are in tandem with SEP3-like genes nor could we identify FLC-like genes through BLAST searches, suggesting that FLC-like genes have been lost in the lineage that led to extant Amborella.
To provide additional support for the sister-relationship between FLC and SQUA and between SEP1 and SEP3 clades, we investigated the synteny of these MADS-box gene loci. We used a novel hybrid mode approach in i-ADHoRe 3.0, a program to detect large-scale synteny between genomic regions35. In hybrid mode, i-ADHoRe 3.0 is able to detect ancient duplication events, even in the presence of additional, more recent, whole-genome duplications. In the Vitis genome, regions around VvFLC2 were found to show significant synteny with those around VvSEP2-VvFUL and VvSEP3-VvAP1 (Supplementary Fig. S2). For example, SEP3-FLC tandems are preceded by a SQUAMOSA PROMOTER BINDING PROTEIN-like gene (SBP) from one monophyletic lineage, while the SEP1-SQUA tandems are preceded by SBP genes from a monophyletic sister group (Fig. 1, Supplementary Fig. S3). These results show that the ancestral SEP and SQUA/FLC lineages underwent an apparent joint round of duplication prior to the radiation of the extant angiosperms, which resulted in the SEP1 and SEP3 clades and the SQUA and FLC clades. Based on synteny and phylogenetic relationships, these duplications were derived from the same segmental duplication in the common ancestor of angiosperms. This is consistent with the recently proposed whole-genome duplication shortly before the radiation of extant angiosperms estimated at 192±2 mya (ref. 36).
Jiao et al. suggested that the gene duplication leading to AGL6 and SEP subfamilies was derived from a whole-genome duplication in the common ancestor of extant seed-plants36. If the AGL6-SEP split indeed originated from such a segmental duplication, we would expect that AGL6 is in tandem with another MADS-box gene, similar to the SEP-SQUA/FLC tandems, and that this other subfamily would be sister to the SQUA/FLC clade. While we find that AGL6-like genes form conserved tandem arrangements with members of another MADS-box gene subfamily: the SOC1-like genes, its exact relation with the SEP-SQUA/FLC tandems will require further research (Supplementary Note 1).
Discussion
The origin and functional diversification of developmental control genes is thought to be a major prerequisite for the evolution of complex morphological traits in eukaryotes. We elucidated the origin of three major MADS-box gene subfamilies. These subfamilies have essential roles in floral transition and flower development: the SQUA and SEP subfamilies act as positive regulators of floral transition and flower development, while FLC genes have mainly been shown to act as vernalization-responsive floral repressors in eudicots.
The concept of vernalization originates from the early observation that winter varieties of cereals require prolonged cold to flower, while spring varieties flower soon after sowing12. Using a combined approach of synteny and phylogeny reconstruction, we were able to unambiguously identify FLC-like genes in monocots. This raises the question whether the findings for FLC in Arabidopsis can be translated to cereal crops and how FLC function diverged in flowering plants. The absence of FLC genes in monocots has previously been used as a major argument to claim an independent origin of vernalization response in these taxa12,13,14. In the future, it will be interesting to see to what extent functional data from Arabidopsis on FLC can be translated to its relatives in temperate cereals or vice versa and whether this can advance our understanding on the vernalization pathway in both cereals and core eudicots. Besides this, FLC genes appear to have been lost in several lineages of flowering plants (Supplementary Fig. S4). It will be interesting to study the frequency and taxonomic distribution of these gene losses and how the floral transition, especially temperature response, evolved in the absence of FLC-like genes in these species.
The results of functional analyses of some members of the OsMADS51 clade (Supplementary Fig. S1), and also our expression data, suggest that some members of the FLC subfamily are controlled by vernalization in temperate grasses, similar to FLC in Arabidopsis. Whether this regulation by cold temperatures is conserved between eudicots and temperate grasses remains to be determined. Also the conservation of underlying gene-regulatory networks and downstream functions of FLC-like genes requires additional research. It is tempting to speculate that some aspects of FLC regulation and functions are conserved throughout angiosperms. Even more intriguingly, some members of the SQUA subfamily, which is sister to the FLC clade, are also regulated by vernalization, although in a positive manner. Examples include VERNALIZATION1 (VRN1) in the vernalization-sensitive grass species Triticum aestivum14,37 or APETALA1 and FRUITFULL in Arabidopsis thaliana whose transcript levels change in response to cold38. However, as also members of other MADS-box gene subfamilies, such as STMADS11-like genes in grasses39, are controlled by vernalization, this type of temperature-dependent regulation may have evolved multiple times independently.
The conserved SEP3-FLC and SEP1-SQUA tandem arrangements in core eudicot and monocot genomes, in combination with the phylogenetic relationships, indicate that the ancestors of these subfamilies originated from a single tandem duplication (Fig. 4). The exact timing of the tandem duplication, as well as the relationship with AGL6-SOC1 tandems, awaits additional combined synteny/phylogeny studies, including more genomes from gymnosperms, as well as basal angiosperms. It is, however, clear that before the diversification of angiosperms, a large-scale duplication, whole-genome or segmental, resulted in the duplication of the ancestral SEP-SQUA/FLC tandem, which gave rise to the SEP3, SEP1, SQUA and FLC gene clades in a concerted manner.
(a) Tree summarizing our current understanding of the phylogenetic relationships based on our and previous analyses. The grey diamond and grey stars denote the inferred tandem duplication (TD) and angiosperm whole-genome duplication (WGD) respectively. (b) Evolutionary scenario explaining how a single-tandem duplication and subsequent large-scale duplications expanded the SEP, AGL6, SQUA and FLC lineages.
Clarifying the origin and evolution of plant MADS-box gene clades is of strong interest for evolutionary developmental biologists, as it helps to trace the evolution of developmental gene-regulatory networks in plants. Bringing our findings into a larger context, the origin of major flowering MADS-box gene subfamilies by tandem duplication and subsequent segmental duplications may be linked to the elaboration of the reproductive transition process and of reproductive meristem identities. The evolutionary origin of flowering plant meristems with specific inflorescence and floral ‘identities’ is still poorly understood, and homologies of reproductive meristem identities between gymnosperms and angiosperms are debated40. The origin of distinct inflorescence and floral meristems may be associated with the evolution of SQUA and SEP gene functions. These genes are essential for the synorganization of reproductive meristems and primordia in the bisexual flower of angiosperms. The potential loss of these genes in gymnosperms may have had consequences for the evolution of meristem identities in these plant lineages. The investigation of gymnosperm genomes, such as the spruce genome41, could provide further insights into the evolution of these enigmatic genes.
Methods
Local synteny
Local synteny was qualitatively determined using the genome browsers implemented in Phytozome42, PLAZA 2.5 (ref. 43), the Sol Genomics Network44 and the Amborella Genome Project ( http://www.amborella.org; funded by the National Science Foundation grant #0922742). Tandem repeats and shared syntenic markers around SEPALLATA- and SQUAMOSA-like genes were identified by BLAST searches against the GenBank database.
Detecting synteny using i-ADHoRe
First, a data set was composed consisting of all angiosperm proteins from PLAZA 2.5 (ref. 43), combined with proteins of more recently sequenced species, Brassica rapa45, tomato46 and potato47. Additionally the Vitis vinifera annotation was downloaded from PLAZA 2.5 and converted to input for i-ADHoRe 3.0 (ref. 35). An all-against-all blastp was run (version 2.2.27+ using default settings)48 to determine pairwise similarities between all proteins in the data set. Next, using tribe-MCL49, the blast output was clustered into homologous gene families, with relaxed settings so even ancient homologues are included in the same gene family (settings: blast-m9, blast-ecut=1e-03, blast-score=e, mcl-I=1.2 and mcl-scheme=4).
For the detection of significant synteny in grapevine, i-ADHoRe 3.0 (ref. 35) was used in hybrid mode, in this mode first colinear regions are detected and hidden from the data set and in a next step the remaining fraction of the genome is scanned for additional syntenic regions.
(cluster_type=hybrid, cloud_gap_size=10, cloud_cluster_gap=15, cloud_filter_method=binomial_corr, gap_size=30, cluster_gap=35, q_value=0.75, alignment_method=gg2, level_2_only=false, prob_cutoff=0.001, anchor_points=3 and multiple_hypothesis_correction=FDR). A less stringent run was performed with a prob_cutoff of 0.05, cloud_gap_size=20 and cloud_cluster_gap=25.
Detection of markers without synteny
Vitis MADS box genes (Supplementary Table S1) were extracted together with 40 protein-coding genes up- and downstream and stored as a list per gene. Redundancy due to tandems was removed and the remaining lists were screened for marker genes that could be used to provide additional insight in the origin of the SEP3-FLC, SEP1-SQUA and AGL6-SOC1 tandem arrangements. Here valid markers are sets of at least three homologous genes, which occur in proximity of all three classes MADS-box genes.
Phylogenetic analyses
We performed BLAST searches using core eudicot FLC sequences represented in a published FLC phylogeny11 against the GenBank, TIGR (The Institute for Genomic Research) and AAGP (Ancestral Angiosperm Genome Project) databases to retrieve additional core eudicot FLC-like genes. Based on the observation of conserved tandem repeat arrangements in core eudicot genomes between SEP3-, SEP1-, SQUA- and FLC-like genes, we found OsMADS37 as a candidate FLC-like sequence in rice. This sequence was used in BLASTN searches to identify similar sequences present in the Genbank database and all sequences retrieved were included in a data matrix for phylogenetic analysis. In addition, we putatively identified the ODDSOC2 clade from the Barley (Hordeum) EST collection through weaker similarity with OsMADS37. Using BLAST searches similar sequences were included in further analyses. We attempted to comprehensively sample all major subfamilies of MIKCc-type MADS-box genes50 to investigate the evolutionary affinities of the sequences found. These subfamilies were consistently represented by sequences of at least one asterid, rosid, magnoliophyte, monocot and gymnosperm when available. Finally, known charophyte, moss and fern MIKCc-type MADS-box genes were included. Using the above sampling rationale we obtained a nucleotide data-matrix consisting of 254 sequences, which was aligned using MAFFT v6 (ref. 51) and manually refined using MacClade4 (ref. 52). The sequences used in phylogenetic analyses are listed in Supplementary Table S3.
C-terminal sequences could not be unambiguously aligned and were therefore excluded from the alignment. In addition, several gene specific insertions were also removed which resulted in a final alignment of 528 bp. The jModeltest program53 was used to determine the best-fit model of nucleotide substitution according to the Akaike information criterion, which selected the GTR+I+G evolutionary model.
The maximum-likelihood phylogenetic analysis was performed using PhyML 3.0 (ref. 54). Bootstrap values summarize 100 bootstrap replicates. Bayesian analysis was carried out using MrBayes 3.2 (ref. 55). Two independent runs with each 4 Markov Chain Monte Carlo chains were run for 15,000,000 generations and sampled every 1,000 generations. After convergence indicated by a s.d. of split frequencies <0.02, we removed the first 25% of the sampled trees as burn-in. The posterior distribution over trees is presented as a majority-rule consensus tree and posterior probabilities are indicated at their respective nodes. Both trees were rooted using charophyte MIKCc type MADS-box genes.
qRT–PCR of FLC-like genes in Brachypodium
Brachypodium distachyon plants were grown in pots containing 50:50 soil:vermiculite. Plantlets were pregrown under long-days (16 h light-8 h dark; 54 photons μmol m−2 s−1) at 28 °C until the third leaf fully emerged. These ‘three-leaf’ plantlets were subsequently transferred to another growth chamber at 4 °C (vernalization treatment) or 28 °C (control) (16 h light-8 h dark; 20 photons μmol m−2 s−1) during 6 weeks and plants were harvested prevernalization and at 2, 4, and 6 weeks. Immediately after harvesting, samples were flash frozen in liquid nitrogen. RNA was extracted from whole plants with their root removed using Trizol (Invitrogen, Carlsbad, USA). Subsequently all RNA samples were DNase treated using TURBO DNA-free (Ambion, Austin, USA). Complementary DNA was prepared by reverse transcription using AMV reverse transcriptase (Promega, Madison, USA). qRT–PCR was performed on a StepOne Plus apparatus (Applied Biosystems, Foster City, USA) using Fast SYBR Green Master Mix (Applied Biosystems, Foster City, USA). The ubiquitin-conjugating enzyme 18 gene (UBC18; Bradi4g00660) was used as a reference gene to normalize the samples56. Relative gene expression change was calculated using the delta-delta Ct method. Error bars represent the s.e. of three biological replicates, which are the mean of three technical replicates. The following primers were used for qRT–PCR: Bradi2g59187 (F: 5′-AAATCCAAGATATTGGCAAAACG-3′, R: 5′-CCTTAGGCTCACTGGAGTTCTCA-3′), Bradi2g59120 (F: 5′-CCGGCAAGCTCTACGAGTACTC-3′, R: 5′-GCTCCCGCAAATTGCTGAT-3′, Bradi3g41297 (F: 5′-CAATCTGAGGATGAAGGTGTCACA-3′, R: 5′-GCTTGACAAGTTGTTCGCTTTCT-3′) and UBC18 (F: 5′-GTCGACTTCCCCGAGCATTA-3′, R: 5′-ATAGGCGCCGGGTTGAG-3′).
Additional information
How to cite this article: Ruelens, P. et al. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat. Commun. 4:2280 doi: 10.1038/ncomms3280 (2013).
References
Anderson, J. T., Willis, J. H. & Mitchell-Olds, T. Evolutionary genetics of plant adaptation. Trends Genet. 27, 258–266 (2011).
Amasino, R. M. & Michaels, S. D. The timing of flowering. Plant. Physiol. 154, 516–520 (2010).
Michaels, S. D. & Amasino, R. M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11, 949–956 (1999).
Adrian, J., Torti, S. & Turck, F. From decision to commitment: the molecular memory of flowering. Mol. Plant 2, 628–642 (2009).
Searle, I. et al. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes. Dev. 20, 898–912 (2006).
Ratcliffe, O. J., Nadzan, G. C., Reuber, T. L. & Riechmann, J. L. Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132 (2001).
Ratcliffe, O. J., Kumimoto, R. W., Wong, B. J. & Riechmann, J. L. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169 (2003).
Kim, D. H. & Sung, S. The Plant Homeo Domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc. Natl Acad. Sci. USA 107, 17029–17034 (2010).
Schranz, M. E., Quijada, P., Sung, S. B., Lukens, L., Amasino, R. & Osborn, T. C. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics 162, 1457–1468 (2002).
Nah, G. & Chen, Z. Tandem duplication of the FLC locus and the origin of a new gene in Arabidopsis related species and their functional implications in allopolyploids. New Phytol. 186, 228–238 (2010).
Reeves, P. A. et al. Evolutionary conservation of the FLOWERING LOCUS C-mediated vernalization response: evidence from the sugar beet (Beta vulgaris). Genetics 176, 295–307 (2007).
Chouard, P. Vernalization and its relations to dormancy. Annu. Rev. Plant Physiol. 11, 191–238 (1960).
Alexandre, C. M. & Hennig, L. FLC or not FLC: the other side of vernalization. J. Exp. Bot. 59, 1127–1135 (2008).
Kim, D. H., Doyle, M. R., Sung, S. & Amasino, R. M. Vernalization: winter and the timing of flowering in plants. Annu. Rev. Cell Dev. Biol. 25, 277–299 (2009).
Hemming, M. N. & Trevaskis, B. Make hay when the sun shines: the role of MADS-box genes in temperature-dependant seasonal flowering responses. Plant Sci. 180, 447–453 (2011).
Rigault, P. et al. A white spruce genecatalog for conifer genome analyses. Plant Physiol. 157, 14–28 (2011).
Melzer, R., Wang, Y. Q. & Theissen, G. The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Semin. Cell Dev. Biol. 21, 118–128 (2010).
Veron, A. S., Kaufmann, K. & Bornberg-Bauer, E. Evidence of interaction network evolution by whole-genome duplications: a case study in MADS-box proteins. Mol. Biol. Evol 24, 670–678 (2007).
Shan, H. et al. Evolution of plant MADS box transcription factors: evidence for shifts in selection associated with early angiosperm diversification and concerted gene duplications. Mol. Biol. Evol. 26, 2229–2244 (2009).
Jiao, Y. et al. A genome triplication associated with early diversification of the core eudicots. Genome. Biol. 13, R3 (2012).
Vekemans, D. et al. Gamma paleohexaploidy in the stem-lineage of core eudicots: significance for MADS-box gene and species diversification. Mol. Biol. Evol. 29, 3793–3806 (2012).
Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E. M. & Smyth, D. R. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development 119, 721–743 (1993).
Pelaz, S., Ditta, G. S., Baumann, E., Wisman, E. & Yanofsky, M. F. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405, 200–203 (2000).
Kaufmann, K. et al. Target genes of the MADS transcription factor SEPALLATA3: integration of developmental and hormonal pathways in the Arabidopsis flower. PLoS Biol. 7, e1000090 (2009).
Tang, H., Bowers, J. E., Wang, X. & Paterson, A. H. Angiosperm genome comparisons reveal early polyploidy in the monocot lineage. Proc. Natl Acad. Sci. USA 107, 472–477 (2010).
Causier, B., Castillo, R., Xue, Y., Schwarz-Sommer, Z. & Davies, B. Tracing the evolution of the floral homeotic B- and C-function genes through genome synteny. Mol. Biol. Evol. 27, 2651–2664 (2010).
Jaillon, O. et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467 (2007).
Zahn, L. M. et al. The evolution of the SEPALLATA subfamily of MADS-box genes: a preangiosperm origin with multiple duplications throughout angiosperm history. Genetics 169, 2209–2223 (2005).
Kim, S. L., Lee, S., Kim, H. J., Nam, H. G. & An, G. OsMADS51 is a short-day flowering promoter that functions upstream of Ehd1, OsMADS14, and Hd3a. Plant Physiol. 145, 1484–1494 (2007).
Arora, R. et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8, 242 (2007).
Kapazoglou, A. et al. The study of two barley Type I-like MADS-box genes as potential targets of epigenetic regulation during seed development. BMC. Plant. Biol. 12, 166 (2012).
International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 463, 763–768 (2010).
Greenup, A. G. et al. ODDSOC2 is a MADS box floral repressor that is down-regulated by vernalization in temperate cereals. Plant Physiol. 153, 1062–1073 (2010).
Winfield, M. O., Lu, C., Wilson, I. D., Coghill, J. A. & Edwards, K. J. Cold- and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC. Plant Biol. 9, 55 (2009).
Proost, S. et al. i-ADHoRe 3.0—fast and sensitive detection of genomic homology in extremely large data sets. Nucleic Acids Res. 40, (2012).
Jiao, Y. et al. Ancestralpolyploidy in seed plants and angiosperms. Nature 473, 97–100 (2011).
Yan, L. et al. Positional cloning of the wheat vernalization gene VRN1. Proc. Natl Acad. Sci. USA 100, 6263–6268 (2003).
Hannah, M. A., Heyer, A. G. & Hincha, D. K. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 1, e26 (2005).
Kane, N. A. et al. TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant. Physiol. 138, 2354–2363 (2005).
Rudall, P. J. & Bateman, R. M. Defining the limits of flowers: the challenge of distinguishing between the evolutionary products of simple versus compound strobili. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 365, 397–409 (2010).
Nystedt, B. et al. The Norway spruce genome sequence and conifer genome evolution. Nature 497, 579–584 (2013).
Goodstein, D. M. et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186 (2012).
Van Bel, M. et al. Dissecting plant genomes with the PLAZA comparative genomics platform. Plant Phys. 158, 590–600 (2012).
Bombarely, A. et al. The Sol Genomics Network (solgenomics.net): growing tomatoes using Perl. Nucleic Acids Res. 39, D1149–D1155 (2010).
Wang, X. et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039 (2011).
The Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641 (2012).
The Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 475, 189–195 (2011).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Enright, A. J., Van Dongen, S. & Ouzounis, C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002).
Becker, A. & Theissen, G. The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29, 464–489 (2003).
Katoh, K. & Toh, H. Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics 9, 212 (2008).
Maddison, W. P. & Maddison, D. R. MacClade 4: analysis of phylogeny and character evolution Sinauer Associates 4.06 (2003).
Posada, D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008).
Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML3.0. Syst. Biol. 59, 307–321 (2010).
Ronquist, F. & Huelsenbeck, J. P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003).
Hong, S. Y., Seo, P. J., Yang, M. S., Xiang, F. & Park, C. M. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC. Plant Biol. 8, 112 (2008).
Acknowledgements
We thank Eric Schranz for comments on a previous version of the manuscript. Thanks to Steven Janssens for helpful tips and suggestions on the phylogenetic analyses. Thanks to Niklas Dochy for assistance during qRT–PCR. K.K. wishes to thank the Alexander-von-Humboldt-Foundation and the BMBF for funding. K.G. and P.R. are supported by FWO grants G.0607.11N and G.0657.13N and P.R. is supported by an IWT fellowship. G.T. thanks the FSU for continuous support.
Author information
Authors and Affiliations
Contributions
K.K. and K.G. function as cosenior authors. The study was designed by K.K. and K.G. and started in the lab of G.T. Genomic tandem arrangements were observed by K.K., RAdM and P.R. Phylogenetic reconstructions were performed by P.R. and K.G. S.P. detected syntenic regions. qRT–PCR experiments were set-up by P.R. and K.G. The paper was written by K.K., K.G., and P.R. with input from G.T. and S.P.
Corresponding author
Ethics declarations
Competing interests
K.K., K.G. and P.R. are authors on a patent application entitled 'Methods and means for modulating flowering time in monocot plants', patent application number EP13174566. The remaining authors declare no conflicts of interest.
Supplementary information
Supplementary Information
Supplementary Figures S1-S7, Supplementary Tables S1-S3, Supplementary Note 1 and Supplementary References (PDF 3787 kb)
Rights and permissions
About this article
Cite this article
Ruelens, P., de Maagd, R., Proost, S. et al. FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4, 2280 (2013). https://doi.org/10.1038/ncomms3280
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms3280
This article is cited by
-
Reciprocal expression of MADS-box genes and DNA methylation reconfiguration initiate bisexual cones in spruce
Communications Biology (2024)
-
Integrated transcriptome and proteome analysis provides insights into CpFPA1 for floral induction in Chimonanthus praecox (Magnoliidae) without FLC in genome
Plant Cell Reports (2024)
-
Genome-wide identification and analysis of MIKC-type MADS-box genes expression in Chimonanthus salicifolius
Genes & Genomics (2023)
-
Diploid and tetraploid genomes of Acorus and the evolution of monocots
Nature Communications (2023)
-
Glucose-driven TOR–FIE–PRC2 signalling controls plant development
Nature (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.