Abstract
DNA double-strand break repair is essential for maintenance of genome stability. Recent work has implicated a host of chromatin regulators in the DNA-damage response, and although several functional roles have been defined, the mechanisms that control their recruitment to DNA lesions remain unclear. Here we find that efficient double-strand break recruitment of the INO80, SWR-C, NuA4, SWI/SNF and RSC enzymes is inhibited by the non-homologous end-joining machinery, and that their recruitment is controlled by early steps of homologous recombination. Strikingly, we find no significant role for H2A.X phosphorylation in the recruitment of chromatin regulators, but rather their recruitment coincides with reduced levels of H2A.X phosphorylation. Our work indicates that cell cycle position has a key role in DNA repair pathway choice and that recruitment of chromatin regulators is tightly coupled to homologous recombination.
Similar content being viewed by others
Introduction
Cell viability and genomic stability are frequently threatened by chromosomal DNA double-strand breaks (DSBs). DSBs can be induced by endogenous free oxygen radicals, collapsed replication forks or exposure to DNA-damaging agents, such as ionizing radiation, ultraviolet light and chemicals1. The failure or improper repair of DSBs can result in cell death or gross chromosomal changes, including deletions, translocations and fusions that promote genome instability and tumorigenesis2. Consequently, cells have developed complex signalling networks that sense DSBs, arrest the cell cycle and activate the repair pathways.
Eukaryotic cells have evolved two major mechanisms that repair chromosomal DSBs, non-homologous end joining (NHEJ) and homologous recombination (HR). NHEJ is the predominant DSB repair mechanism in the G1 phase of the cell cycle, whereas HR predominates in the S and G2 phases3,4,5,6,7. In the case of NHEJ, the broken DNA ends are recognized and bound by the Ku70/Ku80 heterodimer, which subsequently recruits other factors to facilitate ligation of the ends8,9,10. In contrast, DSB repair by HR relies on sequence homology from an undamaged sister chromatid or a homologous DNA sequence to use as a template for copying the missing information. The first step of HR involves extensive processing of the DSB such that the 5′ ends of the DNA duplex that flank the DSB are resected to generate long, 3′ single-stranded tails11. Notably, extensive processing of the DSB ends is inhibited in G1 phase cells by the Ku70/80 complex7, and increased CDK activity at the G1/S boundary activates DSB processing during later cell cycle phases4,5,12.
DSB processing regulates the differential recruitment of two functionally related, checkpoint kinases ATM and ATR (Tel1 and Mec1, respectively in budding yeast). ATM recruitment does not require extensive DSB processing, whereas recruitment of the ATR/ATRIP (scMec1/Ddc2) checkpoint kinase complex requires the binding of the single-stranded binding protein replication protein A (RPA) to the processed DNA13,14. One of the most intensively studied targets for checkpoint kinases is the histone variant H2A.X, which is phosphorylated at a C-terminal serine residue (H2A S129 in yeast or H2A.X S139 in higher eukaryotes; termed H2A.X phosphorylation (γH2AX)). The formation of γH2AX is one of the earliest events at a DSB, and this mark spreads over at least a megabase of chromatin adjacent to each DSB in mammalian cells, and up to 50 kb on each side of a DSB in budding yeast15,16. Although γH2AX is not essential for the initial recruitment of DSB response factors, it has a role in stabilizing the binding of checkpoint factors to DSB chromatin17. Besides its role in the DNA-damage checkpoint, γH2AX has also been proposed to recruit chromatin-regulatory factors, namely the ATP-dependent chromatin remodelling complexes INO80 and SWR-C18,19. These results have established γH2AX as both a ubiquitous hallmark and regulator of the chromatin response to DSBs.
In budding yeast, the DSB recruitment of chromatin regulators has been monitored primarily in asynchronous cell populations, and thus it is unclear if these events are linked to NHEJ or HR. To investigate whether the chromatin response to DNA damage is defined by a specific DSB repair pathway, we induced a single DSB within yeast cells synchronized in either G1 or G2/M cell cycle phases, and chromatin immunoprecipitation (ChIP) assays were performed to follow recruitment of many chromatin regulators. We surprisingly find that subunits of the INO80, SWR-C, NuA4, SWI/SNF and RSC enzymes are primarily recruited outside of G1 phase, with the key NHEJ factor Ku70 inhibiting the recruitment of each of these enzymes in G1 cells. Furthermore, we find that recruitment of all chromatin regulators requires DSB processing and the Rad51 recombinase. In contrast to previous reports, we find that γH2AX has no significant role in the recruitment of chromatin regulators to DSBs in either G2/M or asynchronous cells, though our data do suggest that chromatin regulators may enhance γH2AX dynamics during the HR process.
Results
Recruitment of chromatin regulators is cell cycle regulated
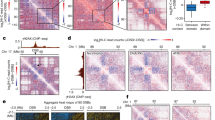
We use an established yeast system that has proven invaluable for monitoring the DSB recruitment of repair factors and chromatin regulators by ChIP analyses. This system allows for a single, persistent DSB to be induced on chromosome III by galactose-dependent expression of the HO endonuclease in a yeast strain that lacks homologous donor sequences20 (hmlΔhmrΔ; Fig. 1a). To investigate whether recruitment of chromatin regulators might be linked to the NHEJ or HR repair pathways, cells were first synchronized in G1 phase with alpha factor (αF) mating pheromone, and then released into three different media conditions: (1) galactose and αF, to induce a DSB in G1 cells; (2) galactose and hydroxyurea, to induce a DSB, as cells exit G1 phase and arrest in S phase; and (3) galactose and fresh media, to induce a DSB, as cells exit G1 and subsequently arrest at the G2/M DNA-damage checkpoint. Cell cycle arrest was confirmed by flow-cytometry analysis (Supplementary Fig. S1a). In this initial study, we followed recruitment of the Arp5 subunit of the INO80 chromatin remodelling enzyme. Surprisingly, recruitment of Arp5 was very low in G1 cells and in cells arrested in S phase. In contrast, Arp5 recruitment was robust in cells that had received a DSB outside of G1 phase and accumulated at the G2/M cell cycle checkpoint (Fig. 1b). To further investigate these results, cells were arrested in either G1 phase with αF or in G2/M with nocodazole, followed by galactose addition to induce a DSB. Initial cell cycle arrest was confirmed by flow cytometry (Supplementary Fig. S2a). Once again, recruitment of INO80, monitored by both Arp5 and the catalytic Ino80 subunit, was robust only when a DSB was induced in G2/M cells, with low levels of recruitment observed at a DSB induced in G1 cells (Fig. 1c and Supplementary Fig. S2c). Consistent with previous findings in asynchronous cultures18,19,21, recruitment of INO80 in G2/M-arrested cultures as well as asynchronous cultures was observed within a 10-kb chromatin domain adjacent to the DSB, and recruitment continued for at least 4 h after DSB formation (Fig. 1c and Supplementary Fig. S2g). Importantly, recruitment of the NHEJ factor yKu70 was also monitored, and in this case DSB recruitment was equal in both G1 and G2/M cells, similar to previous studies7,12 (Supplementary Fig. S2f).
(a) Schematic of chromosome III of a donorless yeast strain harbouring a galactose-inducible HO endonuclease. Primers used during ChIP analyses are indicated according to their distance from the DSB, and designated with a ‘−’ for centrosomal proximal and ‘+’ for centrosomal distal. (b) A wild-type, donorless strain was arrested in G1 using αF and then split into three cultures: maintained in αF-arrest (‘G1’), released into fresh media containing 0.2 M hydroxyurea (‘S(HU)’) or released into fresh media alone (‘G2’). Galactose was also added at this time to induce a single DSB. Arp5 recruitment to areas surrounding the HO cut site was monitored by ChIP. (c,d) A wild-type, donorless strain was arrested with either αF (‘G1’) or nocodazole (‘G2/M’), after which a DSB was induced by addition of galactose for the indicated times. Recruitment of various chromatin remodelling complexes to the DSB region was monitored by ChIP using antibodies to the indicated enzyme subunit. Fold enrichment reflects the %IP values normalized to the ACT1 locus, relative to time zero values. (e) H2A phosphorylation (γH2AX) is cell cycle regulated. Cells were treated as in c, and levels of γH2AX were determined and normalized to levels of histone H3 also determined by ChIP. Data shown represent at least two biological replicates; error bars represent s.e.m.
Given the unanticipated result of differential recruitment of INO80 during the cell cycle, we conducted further ChIP assays to monitor recruitment of several other chromatin regulators, including subunits of the SWR-C, SWI/SNF and RSC remodelling enzymes, as well as the NuA4 histone acetyltransferase complex. Interestingly, the recruitment of each of these chromatin regulators was much more robust outside of G1 phase, compared with G1-arrested cells (Fig. 1d and Supplementary Fig. S2d). These data suggest that there may be a common, cell cycle-regulated mechanism for recruitment of multiple chromatin regulators to a DSB.
γH2AX is dispensable for recruitment of chromatin regulators
Previous ChIP studies have indicated that formation of γH2AX is required for efficient DSB recruitment of INO80 and SWR-C within asynchronous cell populations18,19. To understand how this mechanism interfaces with cell cycle regulation, we monitored the levels of γH2AX in chromatin surrounding DSBs formed in our experiments. Surprisingly, the levels of γH2AX surrounding the DSB were much lower in cells outside of G1 compared with those arrested in G1 (Fig. 1e and Supplementary Figs S1b and S3a). These contrasting levels of γH2AX are not due to changes in nucleosome density, as levels of H3 and H2B were reduced only approximately twofold in G2/M samples compared with G1, presumably due to DSB processing (Supplementary Fig. S3d, see below). Levels of γH2AX were also reduced in G2/M samples at early time points after DSB induction, when end processing has not progressed significantly (for example, 30′), and when ChIP samples were processed in buffers containing 0.5 M NaCl (Supplementary Fig. S3b,c). Furthermore, we monitored formation of γH2AX following exposure of synchronized cells to the DSB-inducing agent phleomycin and again observed more robust γH2AX formation in G1 cells compared with G2/M cells, indicating that these cell cycle differences are not unique to an HO-induced DSB (Supplementary Fig. S3e,f). The data suggest that γH2AX levels or dynamics may be dramatically altered in chromatin surrounding DSBs formed within G2/M cells. Furthermore, these results imply that the levels of γH2AX and chromatin regulators are anti-correlated, indicating that γH2AX may not be involved in their recruitment.
To re-examine the role of γH2AX in recruitment of chromatin regulators, we monitored recruitment events in two different strains that lack γH2AX: a strain expressing a derivative of H2A (bulk yeast H2A is the equivalent to mammalian H2A.X) where serine 129 has been changed to an alanine residue (hta1,2-S129A)22, and a strain expressing a truncated H2A derivative that removes the final four C-terminal amino acids, including the Mec1/Tel1 phosphorylation site (hta1,2-S129Δ4)23. Importantly, both of these strains exhibited similar sensitivity to the DNA-damaging agent methyl methanesulfonate, as expected from previous studies24 (Supplementary Fig. S4a). Surprisingly, neither H2A-S129A nor H2A-S129Δ4 reduced INO80 recruitment, irrespective of whether a DSB was induced in asynchronous or G2/M-arrested cells (Fig. 2a and Supplementary Fig. S4c). Indeed, recruitment of the Arp5 subunit of INO80 was slightly elevated in the strain expressing the C-terminal H2A truncation (Fig. 2a). Similar results were found for Sth1 (RSC), Eaf1 (NuA4), Eaf3 (NuA4/Rpd3), Swi2 (SWI/SNF) and Yaf9 (NuA4/SWR-C) (Fig. 2b and Supplementary Fig. S4d,e). Interestingly, however, recruitment of the Snf6 subunit of SWI/SNF complex was markedly decreased in the absence the H2A C-terminus, even though its recruitment is not affected by the H2A-S129A substitution (compare Fig. 2b and Supplementary Fig. S4e), implicating other residues within the H2A C-terminus. Why recruitment of the Swi2 and Snf6 subunits of SWI/SNF differentially respond to the H2A C-terminus remains unclear. However, when taken together, the data indicate that γH2AX does not regulate recruitment of chromatin regulators.
(a–c) Isogenic, donorless wild-type (wt), hta1,2-S129A (S129A) and hta1,2-S129Δ4 (S129Δ4) strains were arrested in G2/M using nocodazole and analysed by ChIP for recruitment of the indicated chromatin-modifying enzyme subunits to the DSB region at the indicated time points after DSB induction. Data shown represent at least two biological replicates; error bars represent s.e.m.
Although our hta1,2-S129Δ4 and hta1,2-S129A alleles were created within the same JKM strain background as two previously published studies, our ChIP data are contradictory18,19. We obtained the previously published hta1,2-S129* strain (also a four residue truncation; GA2824 (ref. 18)) and found that this strain shows similar sensitivity to DNA-damaging agents as our hta1,2-129Δ4 strain (Supplementary Fig. S4a). However, strain GA2824 also exhibits an unexpected, severe growth defect in raffinose or lactate media, and liquid cultures arrested growth at low cell densities (for example, OD600=0.4). Flow-cytometry analysis also demonstrates that asynchronous cultures of GA2824 grown in raffinose media accumulate in the G1 phase of the cell cycle, and furthermore, this cell cycle distribution does not change following galactose addition to induce the HO endonuclease (Supplementary Fig. S4b). These growth defects precluded our ability to obtain high-quality, reproducible ChIP data with this strain. Previous studies with the GA2824 strain have also indicated that γH2AX is required for efficient DSB processing18. However, a recent study shows that γH2AX inhibits DSB processing25, and we also observe increased levels of RPA adjacent to a DSB in our hta1,2-S129Δ4 and hta1,2-S129A strains (Supplementary Fig. S4f), consistent with a negative role for γH2AX in DSB processing. Because DSB processing is restricted in G1 cells, and INO80 and SWR-C are also poorly recruited in G1 cells, it seems likely that the aberrant slow growth and G1 accumulation phenotypes of the GA2824 strain were the cause of the previously observed defects in both DSB end processing and chromatin regulator recruitment, rather than a lack of γH2AX18.
Chromatin regulator recruitment requires DNA-end processing
Our results indicate that the cell cycle regulation of the DSB response has a key role in the recruitment of chromatin regulators to the DSB. Recruitment of chromatin factors outside of G1 coincides with the binding of RPA to single-stranded DNA (ssDNA) that is formed by the extensive processing of the DSB by the redundant Sgs1/Dna2 and Exo1 resection pathways5,26,27,28 (Supplementary Fig. S2d). Notably, this relationship is also consistent with the poor recruitment of Arp5 (INO80) to a DSB induced within HU-treated cells (Fig. 1b), as HU activates cell cycle checkpoints that inhibit DSB processing29. To examine the possibility that DSB recruitment of chromatin regulators requires resection, we monitored recruitment in isogenic sgs1Δ, exo1Δ and sgs1Δexo1Δ strains. Strikingly, recruitment of Arp5 (INO80), Snf6 (SWI/SNF), Sth1 (RSC), Eaf3 (NuA4/Rpd3) or Yaf9 (NuA4/SWR-C) was greatly reduced in the sgs1Δexo1Δ double mutant, with significant reductions occurring 500 bp distal to the DSB and reducing to basal levels by 2.5 kb distal (Fig. 3a). Importantly, the sgs1Δexo1Δ strain showed a cell cycle profile identical to the wild-type strain (Supplementary Fig. S5a). Consistent with the functional redundancy of these processing enzymes, only a minor defect in recruitment of INO80 to a DSB was observed in exo1Δ or sgs1Δ single mutants (Supplementary Fig. S5b). Interestingly, γH2AX levels were also increased in the double mutant, most notably distal from the break (Fig. 3b). These results suggest that DSB processing is required for optimal recruitment of multiple chromatin regulators and that their recruitment correlates with decreased γH2AX levels.
Isogenic, donorless wild-type (wt) and exo1Δsgs1Δ strains were grown asynchronously and analysed by ChIP in the region surrounding the DSB for (a) recruitment of the indicated chromatin-modifying enzyme subunits 4 h after induction (dotted line indicates the HO cut site) and (b) levels of γH2AX at the indicated time points after induction. Data shown represent at least two biological replicates; error bars represent s.e.m.
Previous studies have shown that the Ku70/80 heterodimer inhibits DSB processing in G1 cells, limiting DNA-end resection and promoting NHEJ7. In addition, loss of Ku70 allows Rad52- and Rad51-dependent recombination events to occur efficiently in G1-arrested cells30. To test whether the decreased recruitment of chromatin factors in G1 is due to limited DSB processing, an yku70Δ strain was arrested in G1, and ChIP assays were performed at an HO-induced DSB. Strikingly, recruitment of all chromatin regulators was restored to high levels in the G1-arrested yku70Δ cells (Fig. 4a). In contrast, γH2AX levels were reduced in the G1-arrested yku70Δ strain to levels previously seen in G2/M cultures, once again displaying an inverse relationship to the recruitment of chromatin regulators (Fig. 4b). These results suggest that yKu70/80 inhibits recruitment of chromatin regulators in G1 cells, and furthermore, that recruitment is independent of cell cycle position. Notably, inactivation of Ku70 does not restore the recruitment of INO80 and SWI/SNF in the absence of Sgs1 and Exo1 (Supplementary Fig. S5d), strongly supporting the idea that DSB processing facilitates the recruitment of chromatin-regulatory factors at the DSB.
Isogenic, donorless wild-type (wt) and yku70Δ strains were arrested in G1 with αF and analysed by ChIP for (a) recruitment of the indicated chromatin-modifying enzyme subunits and (b) levels of γH2AX in the DSB region at the indicated time points after DSB induction. Data shown represent at least two biological replicates; error bars represent s.e.m.
Previous work has shown that the yeast Mre11/Rad50/Xrs2 (MRX) complex directs processing of the initial ~100 bp of DNA proximal to the DSB28. Indeed, we find high levels of the ssDNA-binding protein RPA proximal to the DSB in sgs1Δexo1Δ cells (Supplementary Fig. S5c), a result consistent with MRX-dependent resection. To test whether MRX-dependent processing might be responsible for the residual recruitment of chromatin regulators observed in the sgs1Δexo1Δ double mutant, Arp5 (INO80) recruitment was monitored in wild-type and mre11Δ cells arrested in G2/M. Note that a sgs1Δexo1Δmre11Δ strain was not constructed due to the expected growth defects of this strain. As shown in Supplementary Fig. S5e, Arp5 (INO80) recruitment was lost from DSB proximal chromatin in the absence of Mre11, and overall levels are similar to those found in G1 cells. Taken together, these data suggest a model in which DSB processing controls the recruitment of chromatin regulators, either through direct interactions with ssDNA or by subsequent events of the HR or the DNA-damage checkpoint signalling pathway.
Rad51 is required for recruitment of chromatin regulators
Following DSB processing, the ssDNA ends are initially bound by the ssDNA-binding protein RPA, which is subsequently replaced by the key recombinase, Rad51 (ref. 31). Therefore, we tested whether Rad51 is key for recruitment of chromatin regulators. Strikingly, recruitment of Arp5 (INO80) was nearly eliminated in the G2/M-arrested rad51Δ strain, with a reduction to the twofold recruitment level seen in G1 cells (Fig. 5a). Furthermore, recruitment of Swi2 (SWI/SNF), Snf6 (SWI/SNF), Sth1 (RSC), Eaf3 (NuA4/Rpd3) and Yaf9 (NuA4/SWR-C) were also nearly abolished (Fig. 5a and Supplementary Fig. S6a). In addition, levels of γH2AX were also increased in the absence of Rad51, consistent with one or more chromatin regulators controlling γH2AX dynamics (Fig. 5c). Importantly, Rad51 is not required for DSB processing or establishing the checkpoint response28,31,32 (Supplementary Fig. S6b), indicating that formation of ssDNA is not sufficient for recruitment of chromatin regulators or for decreased levels of γH2AX.
Isogenic, donorless wild-type (wt) and (a) rad51Δ or (b) rad54Δ strains were arrested in G2 with nocodazole and analysed by ChIP for recruitment of the indicated chromatin-modifying enzyme subunits to the region the DSB region at the indicated time points after DSB induction. A dotted line indicates the HO cut site. (c,d) Levels of γH2AX determined by ChIP in the DSB region of experiments described in a and b, respectively. Data shown represent at least two biological replicates; error bars represent s.e.m.
Formation of the Rad51-ssDNA nucleoprotein filament has a key role in the subsequent search and capture of a homologous DNA duplex. Rad51 also recruits Rad54 that is a member of the Snf2/Swi2 family of ATPases and exhibits weak chromatin remodelling activity in vitro33. Rad54 has at least two roles during HR. First, Rad54 has an ATP-independent activity that facilitates Rad51 loading onto DNA proximal to the DSB34,35, and second, Rad54 has an ATP-dependent role to convert the initial joint molecule into a stable, strand-invasion product that can be extended by DNA polymerases35,36. To investigate possible roles for Rad54 in the recruitment of chromatin regulators, a DSB was induced in G2/M-arrested rad54Δ or rad54 K341R strains, the latter of which contains an allele of RAD54 that inactivates its ATPase activity37. ChIP assays for Arp5 (INO80), Swi2 (SWI/SNF), Sth1 (RSC), Eaf3 (NuA4/Rpd3) or Yaf9 (NuA4/SWR-C) indicate a small but reproducible role for Rad54. In all of these cases, there is a defect in recruitment at locations proximal to the DSB, but less of an effect at distal locations (Fig. 5b and Supplementary Fig. S7a). However, very few recruitment defects were observed in the strain harbouring the ATPase-defective version of Rad54 (Supplementary Fig. S7b). In contrast, recruitment of Snf6 (SWI/SNF) was nearly abolished in the absence of Rad54, or when the ATPase activity of Rad54 was inactivated (Supplementary Fig. S7). Thus, recruitment of the Snf6 subunit of SWI/SNF is distinct from both the Swi2 catalytic subunit and other chromatin regulators, requiring both Rad51 and the ATPase activity of Rad54.
Discussion
We have shown here that the recruitment of at least five chromatin-regulatory enzymes—INO80, SWR-C, SWI/SNF, RSC and NuA4—are recruited to a DNA DSB in a cell cycle-dependent manner, with at least five-fold higher levels observed in G2/M cells compared with G1 cells. Our results indicate that recruitment is inhibited in G1 cells by the Ku70/80 complex, and that robust recruitment outside G1 is promoted by early steps of the HR process that lead to formation of the Rad51 nucleoprotein filament. Our data are not inconsistent with roles for chromatin regulators during NHEJ, as recruitment of chromatin regulators is low but not entirely abolished in G1. Indeed, recruitment of the INO80 complex in G1 cells is not affected by loss of Rad51, suggesting an independent mode for recruitment of chromatin regulators at this cell cycle phase. However, Rad51 is at least partially required for recruitment of INO80 in G1 cells that lack Ku70 (Supplementary Fig. S6c). Our data strongly support the view that chromatin regulators primarily impact repair events such as HR that occur following S phase. This idea is consistent with the known roles for the RSC, SWI/SNF, INO80 and SWR-C remodelling enzymes in distinct steps of HR and in cell cycle-checkpoint control22,38,39,40,41.
Although recruitment of human INO80 to DSBs does not require γH2AX42, three studies previously implicated γH2AX in the recruitment of the yeast INO80 and SWR-C remodelling enzymes18,19,21. This conclusion was based primarily on three results: (1) ChIP assays using a strain harbouring an H2A C-terminal truncation allele (hta1,2-S129Δ4); (2) ChIP assays in a mec1 tel1 double mutant; and (3) co-purification of INO80 with γH2AX from cells treated with DNA-damaging agents. Our current data indicate that the interpretation of previous ChIP data was confounded by the cell cycle distribution of the strain used: the previously employed H2A-S129Δ4 strain exhibits an aberrant accumulation of cells in G1, conditions where recruitment of INO80 and SWR-C is poor. Likewise, we envision that the lack of G2 checkpoint arrest in the mec1 tel1 double mutant led to a similar issue. Furthermore, we note that purification of INO80 in low salt buffers leads to co-purification of all four core histones21, so it is expected that some level of γH2AX will be associated with INO80 under DNA-damage conditions. It is perhaps not surprising that γH2AX does not control recruitment of INO80 or SWR-C, because their recruitment requires hours, whereas formation of the γH2AX domain occurs within minutes. In addition, loss of γH2AX leads to relatively little sensitivity to DNA-damaging agents, whereas inactivation of INO80 causes a strong impact on the DNA-damage response18,24. Furthermore, as shown here and previously, the chromatin distribution of γH2AX and INO80 do not coincide at DSBs16,19, and furthermore, our ChIP data show an anti-correlation in the recruitment of chromatin regulators and γH2AX signal. Although it remains a possibility that γH2AX may have a role within G1 cells, our data do not support a dominant role of γH2AX in recruitment of chromatin regulators to DSBs.
Previous studies in budding yeast have demonstrated high levels of γH2AX in both asynchronous cell populations and cells arrested in G1. However, no previous studies have reported γH2AX levels for DSBs induced in cells synchronized outside of G1 phase. We were quite surprised to find a dramatic decrease in γH2AX levels for DSBs induced with G2/M cells. This decrease does not appear to be due solely to DSB processing, as γH2AX levels remain high in rad51Δ cells where DSB resection occurs normally. We envision that γH2AX may be established at normal levels in G2/M cells, but that it is subjected to enhanced dynamics, likely catalysed by one or more chromatin regulators. One possibility is that the low levels of γH2AX reflect dynamic exchange of H2A for H2A.Z by the SWR-C complex22,43,44, although we find that γH2AX levels are not increased in a G2/M-arrested swr1Δ strain (Supplementary Fig. S8). Removal of γH2AX, particularly in G2/M cells, is consistent with recent studies and our own data, indicating a negative impact of γH2AX on DSB processing25 (Supplementary Fig. S4f).
We note that, while all the chromatin-modifying complexes examined share a common set of requirements for recruitment to a DSB, only the Snf6 subunit of SWI/SNF shows a strong requirement for both the H2A C-terminus and Rad54, though neither was needed to recruit the Swi2 catalytic subunit. Our previous study indicated that Snf6 is uniquely associated with SWI/SNF45, so it seems unlikely that it is recruited to DSBs by an independent mechanism. We favour a model in which the Snf6 subunit does not directly associate with nucleosomal DNA, and thus its crosslinking during the ChIP procedure is highly sensitive to small changes in SWI/SNF chromatin interactions.
How might Rad51 control the recruitment of chromatin regulators? One simple possibility is that the Rad51-ssDNA filament functions as an assembly platform for chromatin regulators and that each of these enzymes may directly interact with Rad51. Although there are no subunits held in common among all of the chromatin regulators that we have monitored, we note that each enzyme does harbour a member of the actin-related protein (ARP) family that may provide a common interaction surface46. Alternatively, it is possible that only a limited number of regulators interact directly with Rad51, and the activity of these few enzymes control recruitment of other complexes. Testing this latter possibility may require the development of strains where multiple essential regulators can be removed simultaneously.
Recently, studies in Drosophila and yeast have demonstrated that DSB processing and the Rad51 recombinase are required for long-range, intra-nuclear movements of DSB chromatin during the homology search step of HR40,47, and to regulate repair of heterochromatic DSBs by HR48. Interestingly, yeast studies indicate that the ATPase activity of Rad54 is also essential for enhanced DSB mobility47. Although the INO80 enzyme has been suggested as a candidate factor that catalyses DSB mobility41, our recruitment data suggests that other remodelling enzymes may also contribute, as all enzymes tested require Rad51 for their recruitment to DSBs. How ATP-dependent remodelling might promote chromosome dynamics is currently unclear, though the orchestration of such a complex event may provide one explanation for why so many chromatin regulators are recruited to a DSB.
Methods
Yeast strains
All strains are derivatives of JKM139 or JKM179 (ref. 20) and were generated by the one-step PCR disruption method. Disruptions were confirmed by PCR analysis. Full genotypes are available in Supplementary Table S1. All strains were grown at 30 °C in lactate media (1% yeast extract, 2% bactopeptone, 2% lactic acid, 3% glycerol and 0.05% glucose, pH 6.6) or YPR (1% yeast extract, 2% bactopeptone and 2% raffinose) pre-induction. HO induction was achieved by adding 2% galactose to each culture. Cultures were arrested in G1 using 1 μM αF treatment for 4 h (bar1Δ strains). G2/M arrest was achieved using 30 μg ml−1 nocodazole for 4–5 h. Arrests were confirmed by visual microscopy, followed by budding indices.
Chromatin immunoprecipitation
ChIPs were performed with some modifications as previously described49: 50 ml of mid-log phase cells were cross-linked by adding 1% (final) formaldehyde for 15 min at room temperature, followed by neutralization with 150 mM (final) glycine for 5 min. Cell pellets were washed twice in cold TBS (50 mM Tris-Cl, pH 7.5, and 150 mM NaCl) and then resuspended in cold 400 μl FA-lysis buffer (50 mM HEPES-KOH, pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% sodium deoxycholate) plus 1 × fresh ‘complete’ protease inhibitor cocktail (PIC; Roche). Cells were lysed with an equal volume of glass beads at 4 °C. After glass bead removal, samples were sonicated to shear DNA to an average size of 500 bp. An additional 1 ml FA-lysis buffer plus PIC was added, and the chromatin lysate was purified by centrifugation at 14,000 r.p.m. for a total of 1.5 h at 4 °C.
For most IPs, 100–200 μl of the purified chromatin lysate was diluted up to 400 μl with FA-lysis buffer plus PIC, and 1–2 μl antibody was added. For SWI/SNF IPs, 1% (final) sarkosyl was also added. For high-salt γH2AX IPs, FA-lysis buffer was replaced by FA-500 buffer (50 mM HEPES-KOH, pH 7.5, 500 mM NaCl, 1 mM EDTA, 1% Triton X-100 and 0.1% sodium deoxycholate) All IPs were incubated overnight at 4 °C, followed by incubation with 15 μl equilibrated sepharose protein A beads (50% slurry; Rockland) for 2 h at 4 °C. Pelleted beads were washed at room temperature, for 5–10 min each, sequentially with FA-lysis buffer (except high-salt γH2AX IPs), FA-500 buffer, LiCl wash buffer (10 mM Tris-Cl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP-40 and 0.5% sodium deoxycholate) and Tris-EDTA (10 mM Tris-Cl, pH 7.5, and 1 mM EDTA), followed by elution in Elution buffer (50 mM Tris-Cl, pH 7.5, 10 mM EDTA and 1% SDS) shaking for 10 min at 65 °C. For input samples, 10 μl purified chromatin lysate was diluted in 450 μl Tris-EDTA. All samples (IPs and inputs) were treated with proteinase K (0.2 mg ml−1 final) at 42 °C for 2 h, cross-links reversed by incubation at 65 °C for ≥5 h, and purified by phenol-chloroform extraction and ethanol precipitation. Input samples were diluted 20 × over IP samples during DNA purification.
The immunoprecipitated and input DNA was analysed by quantitative real-time PCR with iTaq SYBR Green Supermix with ROX (Bio-Rad). Primer sequences are available in Supplementary Table S2. Fold enrichment represents the ratio of recovered DNA to input DNA of the break region, normalized to the same ratio obtained for the ACT1 open reading frame. These ratios were additionally normalized to pre-induction (0 h) values and corrected for DSB induction. Percent IP (for anti-RPA only) represents the ratio of immunoprecipitated DNA to input DNA corrected for dilution, and is not normalized to a control region, because those values approached zero. Error bars indicate s.e.m. from at least two independent biological replicas and four PCR reactions.
Western blotting
Whole-cell extracts were prepared by trichloroacetic acid precipitation, and proteins were separated by SDS–polyacrylamide gel electrophoresis in 18% acrylamide gels. Samples were blotted onto polyvinylidene difluoride membranes and probed with antibodies using standard methods.
Antibodies
Rabbit polyclonal antibodies to HA tag (ab9110), Arp5 (ab12099), Yaf9 (ab4468), Eaf3 (ab4467) and H2A-S129phos (γH2AX; ab15083) are commercially available from Abcam. Anti-H2B (39237) is available from Active Motif. Anti-Snf6 and anti-Swi2, anti-RPA, anti-Sth1, anti-Eaf1 and anti-Ku antibodies were kind gifts from J. Reese (Pennsylvania State University), V. Borde (Institut Curie), B. Cairns (University of Utah), J. Cote (Laval University Cancer Research Center) and S.E. Lee (University of Texas Health Science Center at San Antonio), respectively.
Flow cytometry
Approximately 1 ml of mid-log phase (~1–2 × 107) cells were collected per sample, washed in water, fixed in 100% ethanol and incubated at 4 °C rocking overnight. After fixation, cells were again washed in water, resuspended in 50 mM Tris, pH 8.0, containing 200 μg ml−1 RNase A, and incubated at 37 °C for 2–4 h. Samples were then pelleted and resuspended in 50 mM Tris pH 7.5 containing 2 mg ml−1 Proteinase K and incubated at 50 °C for 30–60 min, followed by resuspension in 500 μl FACS buffer (200 mM Tris, pH 7.5, 200 mM NaCl and 78 mM MgCl2). Approximately 100 μl of each sample was then incubated for 10 min at room temperature with 1 ml Sytox solution (50 mM Tris, pH 7.5 and 1 μM Sytox Green (Molecular Probes; S-7020)) and sonicated gently for approximately 30 s directly before analysis on a BD FACSCalibur flow cytometer. Data analysis and preparation was completed with FlowJo.
Additional information
How to cite this article: Bennett, G. et al. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat. Commun. 4:2084 doi: 10.1038/ncomms3084 (2013).
References
Peterson, C. & Côté, J. Cellular machineries for chromosomal DNA repair. Genes Dev. 18, 602–616 (2004).
Khanna, K. K. & Jackson, S. P. DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet. 27, 247–254 (2001).
Moore, J. K. & Haber, J. E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 2164 (1996).
Aylon, Y., Liefshitz, B. & Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 23, 4868–4875 (2004).
Ira, G. et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature 431, 1011–1017 (2004).
Barlow, J. H., Lisby, M. & Rothstein, R. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell 30, 73–85 (2008).
Clerici, M., Mantiero, D., Guerini, I., Lucchini, G. & Longhese, M. P. The Yku70–Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle. EMBO Rep. 9, 810 (2008).
Downs, J. A. & Jackson, S. P. A means to a DNA end: the many roles of Ku. Nat. Rev. Mol. Cell Biol. 5, 367–378 (2004).
Wu, D., Topper, L. M. & Wilson, T. E. Recruitment and dissociation of nonhomologous end joining proteins at a DNA double-strand break in Saccharomyces cerevisiae. Genetics 178, 1237 (2008).
Lieber, M. R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 79, 181–211 (2010).
Bernstein, K. A. & Rothstein, R. At loose ends: resecting a double-strand break. Cell 137, 807–810 (2009).
Zhang, Y., Shim, E. Y., Davis, M. & Lee, S. E. Regulation of repair choice: Cdk1 suppresses recruitment of end joining factors at DNA breaks. DNA Repair 8, 1235–1241 (2009).
Schleker, T., Nagai, S. & Gasser, S. M. Posttranslational modifications of repair factors and histones in the cellular response to stalled replication forks. DNA Repair 8, 1089–1100 (2009).
Polo, S. E. & Jackson, S. P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25, 409–433 (2011).
Rogakou, E. P., Boone, C., Redon, C. & Bonner, W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146, 905–916 (1999).
Shroff, R. et al. Distribution and dynamics of chromatin modification induced by a defined DNA double-strand break. Curr. Biol. 14, 1703–1711 (2004).
Celeste, A. et al. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5, 675–679 (2003).
Van Attikum, H., Fritsch, O., Hohn, B. & Gasser, S. M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119, 777–788 (2004).
Van Attikum, H., Fritsch, O. & Gasser, S. M. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 26, 4113 (2007).
Lee, S. E. et al. Saccharomyces Ku70, Mre11/Rad50, and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell 94, 399–409 (1998).
Morrison, A. J. et al. INO80 and γ-H2AX interaction links atp-dependent chromatin remodeling to dna damage repair. Cell 119, 767–775 (2004).
Papamichos-Chronakis, M., Krebs, J. E. & Peterson, C. L. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 20, 2437–2449 (2006).
Downs, J. A. et al. Binding of chromatin-modifying activities to phosphorylated histone H2A at DNA damage sites. Mol. Cell 16, 979–990 (2004).
Downs, J. a., Lowndes, N. F. & Jackson, S. P. A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408, 1001–1004 (2000).
Chen, X. et al. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature 489, 576–580 (2012).
Gravel, S., Chapman, J. R., Magill, C. & Jackson, S. P. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 22, 2767 (2008).
Mimitou, E. P. & Symington, L. S. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455, 770–774 (2008).
Zhu, Z., Chung, W., Shim, E. Y., Lee, S. E. & Ira, G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134, 981–994 (2008).
Alabert, C., Bianco, J. N. & Pasero, P. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 28, 1131–1141 (2009).
Trovesi, C., Falcettoni, M., Lucchini, G., Clerici, M. & Longhese, M. P. Distinct Cdk1 requirements during single-strand annealing, noncrossover, and crossover recombination. PLoS Genet. 7, e1002263 (2011).
Symington, L. S. & Gautier, J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 45, 247–271 (2011).
Sugawara, N. et al. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature 373, 84–86 (1995).
Ceballos, S. J. & Heyer, W.-D. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim. Biophys. Acta 1809, 509–523 2011.
Wolner, B. et al. Recruitment of the recombinational repair machinery to a DNA double-strand break in yeast. Mol. Cell 12, 221–232 (2003).
Wolner, B. & Peterson, C. L. ATP-dependent and ATP-independent roles for the Rad54 chromatin remodeling enzyme during recombinational repair of a DNA double strand break. J. Biol. Chem. 280, 10855–10860 (2005).
Sugawara, N., Wang, X. & Haber, J. E. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol. Cell 12, 209–219 (2003).
Petukhova, G., Van Komen, S., Vergano, S., Klein, H. & Sung, P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 274, 29453–29462 (1999).
Chai, B., Huang, J., Cairns, B. R. & Laurent, B. C. Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev. 19, 1656–1661 (2005).
Shim, E. Y. et al. RSC mobilizes nucleosomes to improve accessibility of repair machinery to the damaged chromatin. Mol. Cell. Biol. 27, 1602–1613 (2007).
Miné-Hattab, J. & Rothstein, R. Increased chromosome mobility facilitates homology search during recombination. Nat. Cell Biol. 14, 510–517 (2012).
Neumann, F. R. et al. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes Dev. 26, 369–383 (2012).
Kashiwaba, S.-I. & Kitahashi, K. The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochem. Biophys. Res. Commun. 402, 619–625 (2010).
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343 (2004).
Luk, E. et al. Stepwise histone replacement by SWR1 requires dual activation with histone H2A.Z and canonical nucleosome. Cell 143, 725–736 (2010).
Peterson, C. L., Dingwall, A. & Scott, M. P. Five SWI/SNF gene products are components of a large multisubunit complex required for transcriptional enhancement. Proc. Natl Acad. Sci. USA 91, 2905–2908 (1994).
Boyer, L. A. & Peterson, C. L. Actin-related proteins (Arps): conformational switches for chromatin-remodeling machines? Bioessays 22, 666–672 (2000).
Dion, V., Kalck, V., Horigome, C., Towbin, B. D. & Gasser, S. M. Increased mobility of double-strand breaks requires Mec1, Rad9 and the homologous recombination machinery. Nat. Cell Biol. 14, 502–509 (2012).
Chiolo, I. et al. Double-strand breaks in heterochromatin move outside of a dynamic hp1a domain to complete recombinational repair. Cell 144, 732–744 (2011).
Papamichos-Chronakis, M., Petrakis, T., Ktistaki, E., Topalidou, I. & Tzamarias, D. Cti6, a PHD domain protein, bridges the Cyc8-Tup1 corepressor and the SAGA coactivator to overcome repression at GAL1. Mol. Cell 9, 1297–1305 (2002).
Acknowledgements
We thank G. Ira for providing exo1 and sgs1 deletion strains, S. Gasser for providing strain GA2824, J. Cote for providing H2A S129* strain, J. Reese, V. Borde, B. Cairns, J. Cote and S.E. Lee for providing antibodies, and members of the Peterson lab for their helpful discussion. This work was supported by a grant from the NIH (GM54096).
Author information
Authors and Affiliations
Contributions
G.B. designed and performed all experiments, and assisted in manuscript preparation. M.P.-C. and C.L.P. had an equal role in project overview, data interpretation and manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S8 and Supplementary Tables S1-S2 (PDF 1424 kb)
Rights and permissions
About this article
Cite this article
Bennett, G., Papamichos-Chronakis, M. & Peterson, C. DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat Commun 4, 2084 (2013). https://doi.org/10.1038/ncomms3084
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms3084
This article is cited by
-
The NuA4 acetyltransferase and histone H4 acetylation promote replication recovery after topoisomerase I-poisoning
Epigenetics & Chromatin (2019)
-
RNA polymerase II depletion promotes transcription of alternative mRNA species
BMC Molecular Biology (2016)
-
Elucidation of IgH 3′ region regulatory role during class switch recombination via germline deletion
Nature Communications (2015)
-
Mechanisms and principles of homology search during recombination
Nature Reviews Molecular Cell Biology (2014)
-
A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice
Nature Communications (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.