Abstract
The membrane association of the tumour suppressor phosphatase and tensin homologue (PTEN) is required to oppose the phosphatidylinositol-3-kinase/AKT pathway by dephosphorylation of phosphatidylinositol-3,4,5-triphosphate (PIP3). How cytosolic PTEN interacts with its main substrate, PIP3, localized at the inner face of plasma membrane remains unclear. Here we show that PTEN is covalently modified by SUMO1 at both K266 and K254 sites in the C2 domain of PTEN. SUMO1 modification at K266 located in the CBR3 loop, which has a central role in PTEN membrane association, mainly facilitates cooperative binding of PTEN to the plasma membrane by electrostatic interactions. This results in the downregulation of the phosphatidylinositol-3 kinase/AKT pathway and consequently, suppression of anchorage-independent cell proliferation and tumour growth in vivo. Our data demonstrate a molecular mechanism whereby SUMO1 modification is required for PTEN tumour suppressor function by controlling PTEN membrane association and regulation of the phosphatidylinositol-3 kinase/AKT pathway.
Similar content being viewed by others
Introduction
The tumour suppressor gene PTEN (phosphatase and tensin homologue deleted on chromosome 10) is one of the most frequently deleted or mutated in diverse human tumours. PTEN is composed of N-terminal phosphatase domain, C2 domain and C-terminal tail domain (Fig. 1a). PTEN binding to the plasma membrane is a critical regulatory step for PTEN function in antagonizing the phosphatidylinositol-3 kinase (PI3K) signalling pathway by converting phosphatidylinositol 3,4,5-triphosphate (PIP3) to phosphtidylinositol 4,5-biphosphate (PIP2)1,2. PTEN is mainly found in the cytosol and nucleus. Several studies have shown that under normal conditions, only a small fraction of cytosolic PTEN is dynamically bound to the plasma membrane, because PTEN in a constrained conformation has a low affinity for membrane binding2,3. It is still poorly understood how cytosolic PTEN interacts with its main substrate PIP3 localized at the inner face of plasma membrane, although several elements/factors, including the CBR3 loop in the C2 domain4,5,6, N-terminal PIP2-binding motif (Fig. 1a) and PIP26,7, C-terminal tail phosphorylations2,3,8,9,10 and integrity of the phosphatase2,3, have been suggested to be involved in the membrane binding and activation of PTEN.
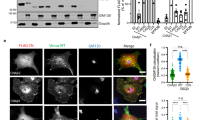
(a) Post-translational modification sites of human PTEN protein. Evolutionarily conserved SUMOylation motif lysines are shown in red. (b) Exogenous PTEN binds to SUMO-conjugating enzyme Ubc9. Lysates from 293T cells transfected with Flag-Ubc9, with or without HA-PTEN were used for immunoprecipitation (IP) with anti-Flag and detected by western blotting. Reciprocally, lysates from 293T cells transfected with HA-PTEN, with or without Flag-Ubc9 were used for IP with anti-HA and detected by western blotting. (c) HeLa cells transfected with or without Flag-Ubc9/His-SUMO1 were lysed and treated with Ni2+-NTA resin for SUMOylation assays11. (d) SENP1+/+ and SENP1−/− MEFs were starved for 24 h and lysed. Lysates as an input were immunoblotted with anti-PTEN and anti-GAPDH antibodies. Lysates were also immunopreciptated with PTEN antibody and then immunoblotted with SUMO1 antibody (with short or long exposure). After stripping, the same membrane was detected with PTEN antibody. Quantification was analysed by ImageJ (V1.45) and the SUMO1–PTEN bands were normalized with the IgG bands. (e) 293T cells co-transfected with PTEN–WT or mutants K254R, K266R and K254R/K266R (KK/RR) with or without Flag-Ubc9/His-SUMO1 were lysed and treated for SUMOylation assays.
PTEN activities and functions are regulated by two levels of transcriptional networks11,12,13 and post-translational modifications, the latter including acetylation, oxidation, S-nitrosylation, phosphorylation, mono- and poly-ubiquitination. Acetylation at K125 and K128 mediated by PCAF (P300/CBP-associated factor) can block PTEN activity14. Ubiquitination at K289 and K13 is regulated by the E3 ubiquitin ligases NEDD4-115,16 and WWP217; the polyubiquitinated PTEN protein undergoes rapid degradation, whereas the monoubiquitinated form manifests the increased translocation to the nucleus. Oxidation of disulphide linkage of catalytic C124 with C71 mediated by reactive oxygen species also negatively regulates PTEN catalytic activity18. S-nitrosylation at the major site C87 (minor sites including C71 and C124) by reactive nitrogen species inactivates its lipid phosphatase activity and mediates PTEN degradation via ubiquitination19. Finally, phosphorylation of PTEN mediated by a series of kinases9,20,21,22,23, such as RhoA-associated kinase on S229/T232 and T319/T321, glycogen synthase kinase 3β on S362/T366 and casein kinase 2 on S370/S380/T382/T383/S385, can regulate protein stability and function in cells. However, SUMOylation of PTEN has not been reported. Here we identify modification SUMO1, with SUMOylation occurring at two sites K266 and K254 in C2 domain of PTEN. Our data indicate that SUMO1 modification at K266 of PTEN is essential for its tumour-suppressor function.
Results
PTEN can be SUMOylated in cells
We analysed the possible SUMOylation sites of PTEN (Supplementary Fig. S1) and found two consensus motifs IK254VE and LK266KD, the former located in Cβ5 and the latter located in CBR3 loop (263KMLKKDK269) of the C2 domain of the PTEN protein4 (Fig. 1a). It has been reported that endogenous PTEN in MCF7 cells associates with Ubc924, which is a sole E2-conjugating enzyme essential for SUMOylation. We have confirmed that exogenously tagged PTEN proteins are also associated with Ubc9 in 293T cells (Fig. 1b). Thus, the protein interaction between PTEN and Ubc9 indicates that PTEN could be SUMOylated in cells. To verify this, we took advantage of a HeLa cell line that contains high levels of endogenous PTEN to perform in vivo SUMOylation assays11. As shown in Fig. 1c, affinity pulldown with Ni2+-NTA resin greatly enriched SUMOylated PTEN with a size of Mr ~75 kDa (the expected normal size of PTEN is 55 kDa), which is PTEN covalently conjugated with one molecule of SUMO1 (SUMO1–PTEN), in HeLa cells transfected with Flag-Ubc9/His-SUMO1, but not in cells without Flag-Ubc9/His-SUMO1. This result suggests PTEN can be SUMOylated in cells.
Highly SUMOylated endogenous PTEN in SENP1−/− MEFs
Among the sentrin/SUMO-specific protease (SENP) protein family, SENP1 is the main de-SUMOylation enzyme for SUMO1-conjugated substrates25,26. We used SENP1−/− mouse embryonic fibroblasts (MEFs) to examine the above hypothesis. Immunoblotting showed that the levels of PTEN protein were comparable in SENP1−/− and SENP1+/+ MEFs. Lysates were also immunoprecipitated with anti-PTEN antibody and then immunoblotted, showing that endogenous SUMO1–PTEN was detected in SENP1−/− and SENP1+/+ MEFs with the same size of 75 kDa by both anti-SUMO1 and anti-PTEN antibodies (Fig. 1d). SUMOylation of PTEN is supposed to be more easily observed in SENP1−/− MEFs than that in SENP1+/+ MEFs. Indeed, we observed that the SUMO1–PTEN in SENP1−/− MEFs was four- to six-fold higher than that in SENP1+/+ MEFs (Fig. 1d). These results demonstrated that endogenous PTEN can be SUMO1-modified cells.
PTEN is modified by SUMO1 at both K266 and K254 sites
To determine the SUMO acceptor sites in PTEN, mutants K254R, K266R or K254R/K266R were generated. We performed SUMOylation assays in 293T cells co-transfected wild-type (WT) PTEN, mutants K254R, K266R or K254R/K266R with or without Flag-Ubc9/His-SUMO1. As expected, one band of SUMOylated Flag-PTEN with a size of Mr ~75 kDa was observed in Ni2+-NTA pulldown of cells transfected with PTEN–WT plasmid (Fig. 1e). The double mutant K254R/K266R completely abolished SUMOylation, whereas the single mutants K254R or K266R greatly reduced SUMOylation in comparison with cells transfected with the WT construct, consistent with the notion that both K254 and K266 are bona fide SUMOylation sites. However, we did not observe a shift in PTEN from 55 to 95 kDa, which is presumably conjugated with two molecules of SUMO1—(SUMO1)2–PTEN (Supplementary Fig. S2), suggesting that K254 and K266 are not simultaneously SUMOylated in cells. Thus, we used only two single (K254R and K266R), but not double, mutants to investigate the physiological function of SUMOylated PTEN. We have confirmed that these two point mutations do not alter other PTEN modifications, such as ubiquitination and acetylation (Supplementary Fig. S3).
PTEN SUMOylation suppresses anchorage-independent growth
To conduct functional analyses, we generated stable PC3luc cell lines by polyclonal lentiviral infections with Lenti-Vector, PTEN–WT, PTEN–K254R or PTEN–K266R. The expression of PTEN was comparable in all clones when assessed by western blotting (Supplementary Fig. S4a), indicating that these two mutations do not affect PTEN transcription rate and protein stability. Additionally, PTEN proteins, including mutants K254R and K266R, still localized predominantly to the cytoplasm (Supplementary Fig. S4b).
To explore whether PTEN SUMOylation affects the transforming potential of each stable PC3luc cell lines, we performed a soft agar colony-forming assay in the presence of 10% fetal bovine serum (FBS). As expected, cells transfected with PTEN–WT showed inhibition of colony growth when compared with the Lenti-Vector-transfected cells. PTEN–K266R-transfected cells did not suppress anchorage-independent growth, whereas PTEN–K254R partially lost the ability to suppress growth. Most importantly, the PTEN–K266R-transfected cells produced numbers and sizes of colonies equivalent to those produced by the Lenti-Vector-transfected cells (Fig. 2a,b). We also observed a similar pattern of results with the soft-agar colony-forming assay in the presence of 1% FBS (Supplementary Fig. S5a). Moreover, we generated stable 293T cells expressing PTEN–WT, PTEN–K254R, PTEN–K266R or Lenti-Vector. The colony-forming assays with these stable cell lines showed the similar pattern of results as those in stable PC3 cell lines (Supplementary Fig. S5b).
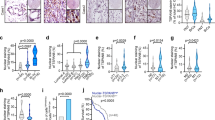
Stable PC3luc transfectants were seeded in 2 ml of medium containing 10% FBS with 0.35% agar at 1×103 cells per well and layered onto the base. The photographs were taken (a), and the number of colonies was scored (b). The same scale bar (500 μm) was used in all images. Each value represents the mean±s.e.m. of three independent experiments with triplicates each. An unpaired (equal variance) t-test was performed on PTEN–WT or PTEN mutants compared with the vector control (***P<0.001), and on PTEN–WT compared with PTEN-K254R (**P<0.01). (c) Tumour growth suppression of PTEN was completely abolished by mutation of K266R. Mice were injected subcutaneously with 2.5×106 PC3Luc cells stably infected with Lenti-Vector, PTEN–WT, PTEN–K254R or PTEN–K266R. Tumour was assessed by bioluminescent imaging with a Xenogen IVIS imaging system at 14 days. (d) Tumour bioluminescent flux was quantified. The data is represented as the mean±s.e.m. of three independent experiments with five mice each. Differences between individual groups as indicated were analysed using the t-test (two-tailed and unpaired), and P-values of <0.01 (**) or <0.001 (***) is considered significant.
PTEN SUMOylation suppresses tumour growth in vivo
To investigate whether PTEN SUMOylation also influences tumour growth in vivo, each stable PC3luc cell line was inoculated subcutaneously into the backs of male severe combined immunodeficiency mice. Bioluminescent imaging assessment was performed at day 14 after injection, showing that no tumours were detected in the PTEN–WT group. Tumours in the PTEN–K254R group grew significantly more slowly than those in both the PTEN–K266R and the Lenti-Vector groups, indicating that K254R mutation results in partial loss of the PTEN tumour suppressor activity. Notably, tumours in the PTEN–K266R group grew as fast as those in the Lenti-Vector group, suggesting that K266R mutation leads to the complete loss of PTEN tumour suppressive activity (Fig. 2c,d). These data suggest that K266 SUMOylation is mainly responsible for PTEN function as a tumour suppressor.
SUMO-site mutations impair PTEN downregulation of p-AKT
As PTEN SUMOylation affects tumour growth in both soft agar colonies and in vivo the mouse xenograft model, we attempted to explore whether the mechanism is involved in regulation of AKT phosphorylation. Lysates from stable PC3luc cells starved for 24 h were immunoblotted, showing that AKT phosphorylation on both T308 and S473 were reduced by 67–69% in PTEN–WT-transfected cells when compared with the Lenti-Vector-transfected cells. In contrast, AKT phosphorylation levels in PTEN–K266R-transfected cells were not significantly different from the Lenti-Vector-infected cells, whereas AKT phosphorylation was reduced by 38–39% in PTEN–K254R-transfected cells when compared with the Lenti-Vector-transfected cells (Fig. 3a,b). The patterns of phospho-AKT were the same as those of findings from both soft-agar colony-forming assay and the in vivo mouse xenograft model described above, demonstrating that K266 SUMOylation of PTEN is required for its capability of tumour suppression through downregulation of phospho-AKT, whereas K254 SUMOylation of PTEN is partially functional. In addition, we have also repeated these experiments in 293T cells stably expressing PTEN–WT, PTEN–K254R, PTEN–K266R or Lenti-Vector (Supplementary Fig. S6), showing the results were very similar to the pattern of results obtained using stable PC3 cell lines.
(a) PC3luc cells stably expressing WT or mutants of PTEN were starved for 24 h and lysed for immunoblotting analysis of AKT phosphorylation at Ser473 and Thr308, AKT, PTEN and GAPDH. (b) Quantitation was analysed by ImageJ 1.45 (NIH, USA). The ratio of the relative intensity of p-AKT(T308) or p-AKT(S473) to total AKT is expressed. The data are presented as the mean±s.d. (n=3). Differences between individual groups as indicated were analysed using the t-test (two-tailed and unpaired), and P-values of <0.01 (**) or <0.001 (***) is considered significant. (c) Haematoxylin and eosin staining of tumour sections from the mouse xenografts at day 32 after injection of PC3luc tumour cells. (d) Tumour sections were immunostained with DAPI (4′,6-diamidino-2-phenylindole (blue)) and antibodies to p-AKT(S473) (green) and PTEN (red). The same scale bar (50 μm) was used in all images. (e) SENP1−/− and SENP1+/+ MEFs stably transduced by Lenti-control-shRNA or Lenti-PTEN-shRNA were starved for 24 h and lysed for immunoblotting analysis of PTEN, p-AKT(T308), p-AKT(S473), AKT and GAPDH. (f) Normalization was performed by the PTEN bands to the GAPDH bands and the p-AKT bands to the AKT bands, respectively. The data are presented as average ±s.d. (n=3). Differences between individual groups as indicated were analysed using the t-test (two-tailed and unpaired), and P-values of <0.01 (**) is considered significant. (g) SENP1−/− and SENP1+/+ MEFs stably transduced by Lenti-control-shRNA or Lenti-PTEN-shRNA were serum-starved for 24 h, then treated with 10% FBS in a time course from 0 to 60 min. Lysates were immunoblotted with anti-p-AKT(T308), anti-p-AKT(S473) and anti-AKT antibodies.
A correlation between tumour growth and AKT phosphorylation controlled by PTEN SUMOylation was observed. We stained sections from the mouse xenografts with haematoxylin and eosin, and observed that the morphology of the three xenograft tumours from the Lenti-Vector, PTEN–K254R and PTEN–K266R groups, was not different, whereas no tumour was observed in the PTEN–WT group (Fig. 3c). For these three xenograft tumour sections, we performed immunofluorescence analysis of PTEN and phospho-AKT. As expected, tumours in the Lenti-Vector group had no detectable endogenous PTEN protein, but high phospho-AKT. However, tumours in the PTEN–K266R group displayed high levels of both PTEN and phosphor-AKT, whereas tumours in the PTEN–K254R group showed high PTEN, but moderate phospho-AKT (Fig. 3d). Thus, these data provide evidence that AKT phosphorylation is relevant to tumorigenesis and controlled by PTEN SUMOylation.
Endogenous SUMO1–PTEN effectively reduces phospho-AKT
We asked if highly SUMOylated PTEN can more effectively reduce phospho-AKT in SENP1−/− MEFs. Indeed, AKT phosphorylation on both T308 and S473 in SENP1−/− MEFs was approximately seven-fold lower than that in SENP1+/+ MEFs (Supplementary Fig. S7). Furthermore, to verify whether PTEN is involved in downregulation of phospho-AKT in the SENP1−/− MEFs system, SENP1−/− and SENP1+/+ MEFs stably transduced by Lenti-control-shRNA or Lenti-PTEN-shRNA for PTEN knockdown were serum-starved for 24 h and lysed for the analysis of phospho-AKT. As expected, AKT phosphorylation on both T308 and S473 in control-shRNA-transduced SENP1−/− MEFs was much lower than those in control-shRNA-transduced SENP1+/+ MEFs. PTEN was effectively reduced by ~90% by PTEN shRNA in both SENP1−/− and SENP1+/+ MEFs. Interestingly, phospho-AKT levels in both SENP1−/− and SENP1+/+ MEFs transduced by PTEN shRNA was almost the same and even higher than those in control-shRNA-transduced SENP1+/+ MEFs (Fig. 3e,f). The MEFs were serum-starved for 24 h, then treated with 10% FBS in a time course from 0 to 60 min for analysis of phospho-AKT. The results showed that phospho-AKT was very low in control-shRNA-transduced SENP1−/− MEFs (Fig. 3g, the second column), whereas phospho-AKT remained high in PTEN-shRNA-transduced SENP1+/+ (Fig. 3g, the third column) and SENP1−/− MEFs (Fig. 3g, the fourth column), in the presence or absence of serum. Phospho-AKT levels were moderate at t=0 and significantly induced by serum in control-shRNA-transduced SENP1+/+ MEFs (Fig. 3g, the first column). These results fully support the hypothesis that SUMO1–PTEN is required for downregulation of AKT phosphorylation.
SUMOylation facilitates PTEN association with the membrane
To explore the underlying mechanism of SUMOylation at K266 of PTEN that is required for the downregulation of AKT phosphorylation and consequent suppression of tumour growth, we constructed the models of PTEN and SUMO1–PTEN for molecular dynamics (MD) simulations. The simulations were based on the X-ray crystal structures of free PTEN (PDB accession code 1D5R, resolution 2.1 Å)4 and SUMO1 (PDB accession code 2IY0, resolution 2.7 Å)25. The structures of PTEN and SUMO1–PTEN (SUMO1 covalently linked to K266 of PTEN) were taken as the starting points for MD simulations. Each MD simulation was carried out using the AMBER suite of programmes (version 8.0) with the parm99 force field (for details, see Methods). On the basis of two equilibrated dynamic trajectories, the snapshots of PTEN and SUMO1–PTEN were respectively extracted and analysed. The results revealed that PTEN in both MD systems are stable and SUMO1 modification is extended from PTEN in the system of SUMO1–PTEN during the simulation time. One eminent feature of charge topology by analysis of the NMR structure of SUMO1 is a stable bipolar surface charge, which on one side displays a positively charged surface and on the other side exhibits a large negatively charged surface27. More recently, it has been reported SUMO1 can bind double-stranded DNA in a sequence-independent manner through electrostatic interaction between SUMO1-positive charge surface and acidic DNA28. It is interesting that SUMO1–PTEN in the short MD simulation show a consensus electropositive interface contributed by K39, K45 and K46 from SUMO1, and by K237, K263 and K313 (Fig. 4a), indicating that SUMO1 modification of PTEN may facilitate cooperative binding of PTEN to the electronegative phospholipid membrane or its substrates by electrostatic interaction.
(a) MD simulations of SUMO1–PTEN binding to membrane by electrostatic interaction. (b) 10 μl per lane of each cytosolic fractions and 50 μl (usually 10 μl, but here 50 μl used for more clearly detected) of membranous protein fractions extracted from SENP1−/− and SENP1+/+ MEFs, respectively, were loaded on SDS–polyacrylamide gel electrophoresis and analysed by western blot. The anti-IGF-1Rβ as membrane marker37 and anti-β-actin as cytosolic marker38 were used for the loading control. (c) Quantitation was analysed by ImageJ 1.45 (NIH, USA), and normalization was performed by the PTEN bands to the IGF-1Rβ bands. The data are expressed as average ±s.d. (n=3). An unpaired (equalvariance) t-test was performed on the relative levels of PTEN in the membrane fraction of SENP1+/+ MEFs compared with those of SENP1−/− MEFs (**P<0.01). (d) Subcellular localizations of PTEN (red) and PIP3 (green) in SENP1−/− and SENP1+/+ MEFs were immunostained and monitored by confocal microscopy. The arrows represent PTEN or PIP3 plasma membrane localization. All images were identically processed and used the same scale bar (50 μm) in all images. (e) The membrane-to-cytosol ratios of PTEN and PIP3 were calculated by ImageJ. A total of 50 staining cells of SENP1−/− or SENP1+/+ MEFs, were used for quantitative analysis.
To confirm the above notion that SUMO1 modification of PTEN directly mediates its membrane association and consequently converting PIP3 to PIP2, we conducted a cellular fractionation assay to show that PTEN in the membrane fraction of SENP1−/− MEFs was four-fold greater than that of SENP1+/+ MEFs (Fig. 4b,c), indicating that PTEN SUMOylation is required for the recruitment of PTEN to the plasma membrane. We also showed that highly SUMOylated PTEN in SENP1−/− MEFs intensely associated with the membrane, ablating plasma membrane PIP3 (Fig. 4d; Supplementary Fig. S8). On the contrary, low SUMOylated PTEN in SENP1+/+ MEFs showed no membrane association, thus leading to PIP3 accumulations of the membrane ruffles. Next, we performed quantitative analysis of 50 cells for each MEFs by ImageJ software, showing that the membrane to cytosol ratios of PTEN proteins were 0.03±0.03 and 0.36±0.09 for SENP1+/+ and SENP1−/− MEFs, respectively. Conversely, the membrane to cytosol ratios of PIP3 were 0.26±0.07 and 0.04±0.05 for SENP1+/+ and SENP1−/− MEFs, respectively (Fig. 4e). As PIP3 is physiologically the major reaction product of PI3K, with PI(4,5)P2 being the major substrate, we used the PI3K inhibitor LY294002 to treat cells. As shown in Supplementary Fig. S9, the immunofluorescence signal of PIP3 staining was significantly reduced, whereas immunofluorescence signal of PTEN staining was not altered in SENP1+/+ and SENP1−/− MEFs. These results support our MD simulation model for PTEN, in which SUMOylation facilitates binding of PTEN to the phospholipid membrane.
K266–SUMO is mainly responsible for PTEN membrane binding
As we have found that PTEN proteins are SUMOylated at K266 and K254, we wanted to know which SUMOylation site is critical for the recruitment of PTEN to the plasma membrane. Thus, we performed the cellular fractionation assays with stable PC3 cell lines. As shown in Fig. 5a, PTEN was highly enriched in the plasma membrane fractions of both PTEN–WT- and PTEN–K254R-transfected cells, although the latter was relatively lower. Surprisingly, PTEN was not recruited to the plasma membrane of PTEN–K266R-transfected cells. We next used a phosphatase assay of diC8-PIP3 with immunoprecipitated PTEN from the same amount of above plasma membrane fractions of each stable cell line. We show that PTEN–WT-transfected cells had the highest phosphatase activity and PTEN–K254R mutant had an intermediate phosphatase activity, whereas the PTEN–K266R mutant and the Lenti-Vector-transfected cells had a basal level of activity (Fig. 5b). This result also accurately reflected the abundance of PTEN in those fractions. Moreover, immunofluorescence staining also confirmed that PIP3 accumulation at the plasma membrane of PTEN–K266R-transfected cells were also significantly increased to the similar levels as those of the Lenti-Vector-transfected cells. However, PTEN–K254R-transfected cells showed low levels of plasma-membrane-bound PIP3 (Fig. 5c). In addition, we have also repeated the cellular fractionation and phosphatase assays in 293T cells transiently transfected with the empty vector, PTEN–WT, PTEN–K254R or PTEN–K266R plasmids, together with SUMO1 and Ubc9, and showed comparable results (Fig. 5d,e) to those of stable PC3 cell lines. Taken together, the above results correlated well with the tumour suppressor phenotype, including mouse xenograft and soft agar colony, and demonstrated that SUMOylation at K266 is mainly responsible for the recruitment of PTEN to the plasma membrane.
(a) A volume of 10 μl per lane of each cytosolic fraction and membranous protein fraction extracted from PC3luc cells stably expressing WT or mutants of PTEN, respectively, was loaded on SDS–polyacrylamide gel electrophoresis and western blotted with anti-PTEN, anti-IGF-1Rβ and anti-β-actin antibodies. (b) Malachite green phosphatase assay using proteins immunoprecipitated with anti-PTEN antibody from equal amounts of membranous fraction extracted from 1×108 of each stable PC3luc cell line expressing WT or mutants of PTEN. The data are presented as average ±s.d. (n=4). Differences between individual groups as indicated were analysed using the t-test (two-tailed and unpaired), and P-values of <0.01 (**) or <0.001 (***) is considered significant. (c) PC3luc cells stably expressing WT or mutants of PTEN were immunostained with anti-PIP3 antibody and monitored by confocal microscopy. The arrows represent PIP3 plasma membrane localization. All images were identically processed and the same scale bar (25 μm) was used in all images. (d) A volume of 10 μl per lane of each cytosolic fraction and membranous protein fraction extracted from 293T cells transiently transfected with the empty vector, Flag-PTEN–WT, –K254R or –K266R together with or without HA-Ubc9 and His-SUMO1, respectively, was loaded on SDS–polyacrylamide gel electrophoresis and western blotted with anti-PTEN, anti-IGF-1Rβ and anti-β-actin antibodies. (e) Malachite green phosphatase assay using proteins immunoprecipitated with anti-PTEN antibody from equal amounts of membranous fraction extracted from 3×107 of each 293T cells transiently transfected with the empty vector, Flag-PTEN–WT, –K254R or –K266R together with or without HA-Ubc9 and His-SUMO1. The data are presented as average ±s.d. (n=4). Differences between individual groups as indicated were analysed using the t-test (two-tailed and unpaired), and P-values of <0.01 (**) or <0.001 (***) is considered significant.
PTEN-controlled tumorigenesis is dependent on K266–SUMO
To investigate the structural basis of the CBR3 loop in PTEN-controlled tumorigenesis, MD simulations, and structural and functional analysis were performed on PTEN and its four mutants. The results show that the conformation of the CBR3 loop could significantly change, owing to sequential multiple replacements of PTEN M-CBR3 (PTEN263AAGAADA269)4. The most greatest deviation of the loop in PTEN263AAGAADA269 is around residue 266 and the positively charged property of the loop has been eliminated, which could lead to a lower ability to bind the membrane4. On the contrary, single mutations at residue 266 had little effect on the perturbation of the CBR3 loop and the whole PTEN conformation (the root-mean-square deviation for the Cα atoms <1.8 Å) in the solvents. These mutations ranged from a similar positively charged residue (PTEN–K266R) to a polar residue (PTEN–K266Q), to a non-polar residue (PTEN–K266A), in comparison with WT PTEN (Fig. 6a). Although the simulation times were limited, the conserved conformations in the PTEN–K266 mutants provide a clue that these various mutants could have the same biological contribution. To verify this, we also generated PC3 cell lines stably expressing PTEN–K266Q, PTEN–K266A or PTEN–G129R, and conducted the soft-agar anchorage-independent growth assays with these mutants. Three point mutants K266R, K266Q or K266A produced soft-agar colonies equivalent in size and number to those produced by the Lenti-Vector- and PTEN–G129R-transfected cells (as a positive control)29 (Fig. 6b,c). Furthermore, we determined the levels of phospho-AKT, showing that PTEN–WT effectively inhibited AKT phosphorylation on both T308 and S473, whereas all other mutants including K266R, K266Q, K266A and PTEN–G129R completely lost this ability (Fig. 6d). Therefore, the tumour formation of PTEN–K266 mutants is ascribed to SUMOylation deficiency instead of a conformational change.
(a) Superimposition of the CBR3 loops isolated from the structures of PTEN and PTEN mutants in both crystal and MD simulations. The sequence alignment of cβ5-CBR3-cβ6 (defined by Lee et al.)4 from the structures is shown in the lower right corner. Residue K266 is highlighted in yellow in both sequences and structures. AAGAADA_5NS (PTEN 263AAGAADA269 at 5-ns MD simulation); WT_5NS (WT PTEN at 5-ns MD simulation); K266R_5NS (PTEN K266R at 5-ns MD simulation); K266Q_5NS (PTEN K266Q at 5-ns MD simulation); K266A_5NS (PTEN K266A at 5-ns MD simulation); CRY (PTEN crystal 1D5R). (b) Stable PC3luc cells stably expressing WT PTEN or mutants (including K266R, K266A, K266Q and G129R) were seeded in 2 ml of medium containing 10% FBS with 0.35% agar at 3×103 cells/ per well and layered onto the base. The photographs of the cells growing in plate and of the colonies developed in soft agar were taken 2 weeks after seeding. All images were identically processed and the same scale bar (500 μm) was used in all images. (c) The number of colonies was scored. Each value represents the mean±s.e.m. of three independent experiments with triplicates each. An unpaired (equalvariance) t-test was performed on Lenti-Vector or PTEN mutants compared with PTEN–WT (***, P<0.001). (d) PC3luc cells stably expressing WT PTEN and mutants were starved for 24 h, and then lysed for immunoblotting analysis of AKT phosphorylation at Ser473 and Thr308, AKT, PTEN and GAPDH.
Furthermore, to more strongly support the hypothesis that the mutation of K266 inhibits PTEN SUMOylation-dependent membrane recruitment, but does not alter the conformation of PTEN thereby hindering its recruitment to the plasma membrane, we made Flag-tagged SUMO1–PTEN (WT, K254R or K266R) fusion expression constructs and generated stable PC3 cell lines by lentiviral infections with these new constructs (Fig. 7a). We conducted western blotting analysis of phospho-AKT (Fig. 7b), the soft-agar colony-forming assays (Fig. 7c) and cellular fraction assays (Fig. 7d). Indeed, all these artificially SUMOylated forms of PTEN were equally recruited to the membrane, although relatively less so when compared with PTEN in the cytoplasmic fractions, which is probably due to the fast dynamic association between PTEN and the inner face of plasma membrane. As expected, similar to PTEN–WT–sumo1, both PTEN–K254R–sumo1 and PTEN–K266R–sumo1 reversed the phenotypic observations of inhibition of phospho-AKT (Fig. 7b) and anchorage-independent growth (Fig. 7c). These results demonstrate that PTEN SUMOylation in PTEN-null PC3 cells is indeed important for PTEN membrane recruitment.
(a) SUMO1–PTEN fusion expression constructs Lenti-PTEN–WT–sumo1, PTEN–K254R–sumo1 and PTEN–K266R–sumo1 were generated (detail see Supplementary Methods) and their structure is shown. PC3luc cells were infected with the prepared pseudovirus containing the lentivirus expression constructs Lenti-PTEN–WT or above SUMO1–PTEN fusion expression constructs. Stably transduced cells selected by puromycin were lysed and immunoblotted with anti-PTEN and anti-β-actin antibodies. Three of SUMO1–PTEN fusion proteins were expressed comparable to the same size of Mr ~70 kDa, whereas the PTEN–WT without fusion of SUMO1 was expressed with a normal size of Mr ~55 kDa. (b) Point mutations of K266R and K254R that impair PTEN function in downregulation of AKT phosphorylation were phenotypically rescued by covalent attachment of SUMO1 to PTEN by gene fusion. PC3luc cells stably expressing Lenti-PTEN–WT–sumo1, PTEN–K254R–sumo1 and PTEN–K266R–sumo1 were starved for 24 h, and then lysed for immunoblotting analysis of AKT phosphorylation at Ser473 and Thr308. (c) The effect of SUMO1–PTEN fusion proteins on anchorage-independent growth was assessed using a soft agar colony assay. PC3luc cells stably expressing Lenti-PTEN–WT–sumo1, PTEN–K254R–sumo1 and PTEN–K266R–sumo1 (including control PC3luc cells with Lenti-Vector) were seeded in 2 ml of medium containing 10% FBS with 0.35% agar at 3×103 cells per well and layered onto the base. The photographs of the cells growing in plate and of the colonies developed in soft agar were taken 2 weeks after seeding. All images were identically processed and the same scale bar (500 μm) was used in all images. (d) A volume of 10 μl per lane of each cytosolic fraction and membranous protein fraction extracted from PC3luc cells stably expressing Lenti-Vector, PTEN–WT–sumo1, PTEN–K254R–sumo1 and PTEN–K266R–sumo1, respectively, was loaded on SDS–polyacrylamide gel electrophoresis and western blotted with anti-PTEN, anti-IGF-1Rβ and anti-β-actin antibodies.
Discussion
Recent studies have revealed that the presence of SUMOylated proteins occurs not only in the nucleus, but also in other cellular compartments, including the cytoplasm, mitochondria, endoplasmic reticulum and the plasma membrane30. PTEN is mainly found in the cytoplasm as a lipid phosphatase that dephosphorylates its substrate at the plasma membrane, but is also found in the nucleus15,16. In this study, we have confirmed that PTEN can be modified by SUMO1 in HeLa cells (Fig. 1c), 293T cells (Fig. 1e) and PC3 cells (Supplementary Fig. S2) using His-tagged SUMO1 conjugates bound to Ni2+-NTA beads. Importantly, we used immunoprecipitation and western blotting to show the high level of endogenous SUMOylated PTEN in SENP1−/− MEFs (Fig. 1d).
In fact, PTEN has also been found to directly translocate to the plasma membrane in certain cell lines and under specific conditions3,20,31. Our finding that SUMO1 modification of PTEN increases PTEN binding to the plasma membrane (Figs 4,5 and 7) may explain why a small fraction of PTEN acts through the dynamic interaction with the inner face of plasma membrane2. Most SUMO targets appear to be SUMOylated to a small percentage at steady state; nevertheless, this low-level SUMOylation causes large effects30. Dynamic cycles of SUMOylation and de-SUMOylation transiently occur within seconds32, and PTEN binds to the plasma membrane for a few hundred milliseconds, which is sufficient to dephosphorylate PIP32. Thus, in view of these time frames, it is reasonable to assume that PTEN SUMOylation and PTEN membrane association can occur in this time.
As previous studies have shown4,5,6, PTEN binds to phospholipid membranes via its C2 domain, in which the CBR3 loop has a central role in the binding. The solvent-exposed loop has four positively charged lysines (K263, K266, K267 and K269) and two hydrophobic residues (M264 and L265). A mutant of this region, PTEN M-CBR3 (263AAGAADA269)4, shows reduced affinity for membranes in vitro compared with WT PTEN. In this study, we found K266 in the CBR3 loop is SUMO modified, and the K266 mutant has been revealed to contribute to increased tumour formation (Fig. 2c). To predict the structural and functional relationship of K266, the MD simulations on PTEN-WT and its mutants, including K266R, K266A and K266Q, were performed and the results showed that the conformations in the PTEN K266 mutants are conserved in 5-ns trajectories (Fig. 6a). Considering the temporal and spatial features of PTEN function, long simulation of PTEN systems in the presence of a negatively charged bilayer can provide more convincing evidences in the mechanism studies of both CBR3 SUMOylation and membrane recruitment. Nevertheless, a series of very short MD simulations in this study have been employed to simply predict potential effects of PTEN mutants; these need to be verified with further biological experiments. However, these various mutants with the same biological contribution have been validated by the soft agar anchorage-independent growth assays (Fig. 6b,c) and the determination of phospho-AKT levels (Fig. 6d). Therefore, PTEN-controlled tumorigenesis is more likely to be K266–SUMO-dependent, but not due to changes in PTEN conformation.
It has been proposed in the PTEN open/closed model that phosphorylation at S370/S380/T382/T383/S385 of the C-terminal tail regulate membrane association3,33. This model is also involved in binding of the C2 domain to phosphatidylserine4 and the PIP2-binding motif to PIP27,34. We have provided evidence that a consensus electropositive interface in SUMO1–PTEN facilitates cooperative binding of PTEN with the electronegative phospholipid membrane or its substrates by electrostatic interaction. Phosphorylation leads to a closed conformation that prevents PTEN association with the plasma membrane9,10; however, if taken into account the electrostatic status, phosphorylation actually supplies negative charges. Therefore, SUMOylation appears to be a positive regulator in controlling PTEN membrane association, whereas phosphorylation is a negative regulator that may neutralize SUMOylation through intramolecular electrostatic interactions. This hypothesis remains to be validated in the future.
We detected enhanced tumour proliferation when K254 was mutated, but this was less than the K266 mutant, which elicits the same tumorigenic phenotype as seen in the PTEN-null condition (Fig. 2). From this observation, it seems that both K266 and K254 are important in regulating tumour formation, but more so when K266 is mutated. According to the principle of electrostatic interaction, the SUMOylation of either K254 or K266 could provide charge to help PTEN in the process of membrane binding. However, K254 is located in the rigid β-strand of the Cβ5 domain and K266 is located in the flexible CBR3 loop; thus, K266 has more potential for conformational changes to induce biological function. This is in agreement with previous studies that the CBR3 loop is necessary for membrane binding. In addition, the SUMO1 protein contains more than 90 residues, and the requirement of space near the modified site is much larger than for other posttranslational modifications (for example, methylation, acetylation, phosphorylation), leading to an unfavourable factor for single SUMOylation of K254 embedded in the Cβ5 domain or dual SUMOylation of adjacent K254 and K266.
In summary, as shown in Fig. 8, our data unravel an unexpected regulatory mechanism that PTEN SUMOylation is required for inhibition of the PTEN/PI3K/AKT pathway. SUMO1 modification of PTEN at K266 mainly facilitates cooperative binding of PTEN to the electronegative phospholipid membrane by electrostatic interaction, then dephosphorylating PIP3 to PIP2, consequently blocking AKT activation and suppressing anchorage-independent cell proliferation and tumour growth in vivo. Our findings might also have important implications in cancer aetiology and cancer therapy, as dysregulation of the PTEN/PI3K/AKT pathway is associated with diverse cancers.
A receptor tyrosine kinase (RTK) activated by its ligand phosphorylates PI3K to convert PIP2 to PIP3. PIP3 serves as a second messenger and together with PDKs activates AKT. Activated AKT, by phosphorylation, mediates the activation and inhibition of several targets, resulting in cell growth, proliferation, survival and transformation. On the other hand, the tumour suppressor PTEN as a lipid phosphatase antagonizes PI3K function and consequently inhibits downstream signalling through Akt. SUMO1 modification at K266 of PTEN mainly facilitates cooperative binding of PTEN to the electronegative phospholipid membrane by electrostatic interaction, then dephosphorylating PIP3 to PIP2, consequently blocking AKT activation and suppressing anchorage-independent cell proliferation and tumour growth in vivo.
Methods
Cell cultures
Human embryonic kidney 293T, 293FT and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS; Hyclone), penicillin and streptomycin (Invitrogen) at 37 °C and 5% CO2. PC3luc expressing a firefly luciferase can be used for living imaging35. PC3luc cells were cultured in RPMI1640 in 10% FCS, penicillin and streptomycin at 37 °C and 5% CO2. MEF cells from SENP1 homozygous null (SENP1−/−) and WT (SENP1+/+) mice36, which was provided by Dr J. K. Cheng in SJTU-SM, were cultured in DMEM with 10% FBS. SENP1+/+ and SENP1−/− MEFs were generated and maintained according to the protocol of Cheng et al.36 Cell transfection was performed using Lipofectamine 2000 (Invitrogen).
Soft agar colony assay
The effect of PTEN and its mutants on anchorage-independent growth was assessed using a soft agar colony assay. Briefly, this assay was performed in six-well plates with a base of 2 ml of medium containing 10 or 1% FBS with 0.6% Bacto agar (Amresco). Stable PC3luc transfectants were seeded in 2 ml of medium containing 10 or 1% FBS with 0.35% agar at 1×103 or 1×104 cells per well and layered onto the base, respectively. The photographs of the cells growing in the plate and of the colonies developed in soft agar were taken, and the number of colonies was scored by ImageJ V1.45 (NIH, USA).
Analysis of SUMO1-modified protein
SUMO1 modification of PTEN was analysed in 293T, HeLa or PC3 cells by the method of His-tagged SUMO1 conjugates binding to Ni2+-NTA beads as previously described11. For analysis of endogenous SUMO1–PTEN, SENP1−/− and SENP1+/+ MEFs were lysed in NEM-RIPA buffer, and 1500 μg of lysates were used for immunoprecipitation with 5 μl of anti-PTEN (26H9) antibody, then immunoblotted with anti-SUMO1 and anti-PTEN antibodies (dilution 1:1000).
Extraction of membrane/cytosol fractions
A total of 8−10×106 cells were used for each extraction, and the extractions were performed according to the manufacturer's instructions of FractionPREP Cell Fractionation kit (BioVision, CA, USA). Both subcellular fractions were further resuspended in 400 μl of buffer, and usually 10 μl per lane of each cytosolic and membranous protein fractions were loaded on SDS–polyacrylamide gel electrophoresis and western blotted. The anti-IGFIRβ as a membrane marker37 and anti-β-actin as a cytosolic marker38 were used for the loading control.
Phosphatase assays
Equal amounts of membranous fractions from 1×108 of each stable PC3luc cell line expressing WT PTEN or mutants were used for immunoprecipitation with 5 μl of anti-PTEN (26H9; Fig. 5b). Equal amounts of membranous fractions from 3×107 of each 293T transiently transfected with the empty vector, Flag-PTEN–WT, –K254R or –K266R, together with or without HA-Ubc9 and His-SUMO1, were used for immunoprecipitation with 5 μl of anti-Flag antibody (Fig. 5e). According to the manufacturer's protocol of Malachite Green Phosphatase Assay Kit (Echelon, Salt Lake City, Utah), the phosphatase reactions with water-soluble diC8-PIP3 (Echelon) were performed with the above immunoprecipitated proteins.
Mouse xenograft models
Murine xenografts were established in 6- to 8-week-old male severe combined immunodeficiency mice using PC3luc cells, which were stably infected with Lenti-Vector, PTEN–WT, PTEN–K254R and PTEN–K266R, respectively. Per injection site, a total volume of 100 μl containing 2.5×106 PC3luc cells were inoculated subcutaneously into the backs. Tumour volume was assessed by imaging isofluorane-anaesthetized mice with the IVIS system (Xenogen, Alameda, CA). Images were obtained 10 min after intraperitoneal injection of 1.5 mg (approx. 75 mg kg−1) D-luciferin (Xenogen) in 100 μl of PBS. The light emitted by luciferase-expressing tumours was quantified using Living Image Version 2.50 (Wavemetrics, Lake Oswego, OR). A heatmap image corresponding to photons/second/cm2/steradian (blue lowest and red highest) was superimposed over the reflected light image of the animals for tumour localization. A rectangular region of interest encompassing the mouse head and body was replicated between images, images were adjusted to identical luminescent scales, and bioluminescence reported as total flux for the region in photons per second. Animals were imaged on day 14 and 21. The data is represented as the mean±s.e.m. of three independent experiments with five mice each. In vivo bioluminescent numerical data accounting for tumour growth was compared between groups using one-way repeated-measures analysis of variance for significance of intergroup comparisons. Statistical analysis was performed using Prism 5 (GraphPad Software, La Jolla, CA). All animal studies were conducted with the approval and guidance of Shanghai Jiao Tong University Medical Animal Ethics Committees.
Immunofluorescene and confocal microscopy
Immunofluorescene staining method was performed as previously described11. Briefly, SENP1−/− and SENP1+/+MEFs were directly seeded into the uncoated 35-mm dishes at a density of 1.0×105 cells. After 24 h, cells were treated with LY294002 (20 μM) or vehicle (DMSO) for 30 min, then washed in PBS once, fixed with 4% freshly prepared formaldehyde in PBS for 8–10 min, and then washed three times with PBS. Cells were permeabilized with 0.1% Saponin (or Triton 100)/PBS for 15 min, blocked in 2.5% normal goat serum in 0.1% Saponin (or Triton 100)/PBS for 30 min–24 h, incubated in the primary antibodies anti-PTEN(138G6, rabbit monoclonal immunoglobulin G, dilution 1:50) and anti-PI(3,4,5)P3 (Z-P345b, mouse monoclonal immunoglobulin G, dilution 1:50) diluted in blocking solution for 1 h, washed three times with blocking solution and then incubated in the second antibody (Alexa 568 anti rabbit, or Alexa 488 anti mouse, dilution 1:500) in blocking solution for 30 min. The cells were then washed twice with blocking solution, and once with PBS. DAPI (4′,6-diamidino-2-phenylindole) was added for DNA staining.
Images were taken with a Zeiss LSM710 Confocal Microscope (Carl Zeiss, Jena, Germany). All confocal images were analysed and quantified using ImageJ v. 1.45 (http://rsb.info.nih.gov/ij/). The membrane-to-cytosol ratio of respective experiments was calculated on single Z-planes using the methods developed by Janetopoulos et al.39 and Ulrich et al.40 For each of MEFs, 50 staining cells were used for quantitative analysis and statistical analysis (Fig. 4d).
Molecular models of PTEN and SUMOylated PTEN
The models of PTEN, SUMOylated PTEN (SUMO1–PTEN) and four mutated PTEN (K266R, K266Q, K266A, M-CBR3--263AAGAADA269) for the MD simulations were constructed based on the X-ray crystal structures of free PTEN (1D5R, resolution 2.1 Å)4 and SUMO1 (2IY0, resolution 2.7 Å)25. The side chains with missing coordinates were reconstructed using the fragment library of the Biopolymer module in Sybyl version 6.8 (Tripos, St Louis, MO). The modified structures were subjected to energy minimization in Sybyl6.8, using the steepest descent method up to the gradient tolerance of 0.05 kcal per mol·Å to relieve possible steric clashes and overlaps of side chains. The three-dimensional structural model of SUMO1–PTEN was built up using the xLeap module in the AMBER suite (version 8.0).
MD simulations
The structures of six models were taken as the starting points for MD simulations. We performed the MD simulation of PTEN–SUMO1 after modelling, to relax the conformation of PTEN–SUMO1 in solution and remove unfavourable contacts arising from the initial rigid-body modelling, whereas other MD simulations on PTEN mutants were used to investigate the structural basis and conformational changes of the CBR3 loop. Each MD simulation was carried out using the AMBER suite of programmes (version 8.0) with the parm99 force field41. Each structure was prepared by using the xLeap module in AMBER, in which protons were added to the structure. All ionizable side chains were maintained in their standard protonation states at pH 7.0. The proteins were solvated in cubic box of water molecules, with a water thickness extending at least 10 Å apart from the protein surface. To avoid the instability that might occur during the MD simulations, the solvated system was subjected to minimization for 5000 cycles with protein restrained and followed by another 5,000 cycles with the whole system relaxed. Then, the system was gradually heated from 0 to 300 K during the first 60 ps by three intervals, followed by equilibrium for 80 ps under constant volume and temperature condition. Afterwards, the system was switched to constant pressure and temperature condition and equilibrated for 100 ps to adjust the system to a correct density. Finally, the production simulations were carried out in the absence of any restraint under constant pressure and temperature condition and two 5-ns MD simulations were then conducted on the PTEN and SUMO1–PTEN to probe the function of SUMOylation. This protocol was applied to all of the simulation systems.
All the MD simulations were performed using the parallel version of PMEMD in AMBER suit. The particle mesh Ewald method was employed to calculate the long-range electrostatic interactions, whereas the lengths of the bonds involving hydrogen atoms were fixed with the SHAKE algorithm42. During the simulations, the integration time step of 2 fs was adopted and structural snapshots were flushed every 500 steps (1 ps). The non-bonded cutoff was set to 10.0 Å, and the non-bonded pair list was updated every 25 steps. Each production simulation was coupled to a 300 K thermal bath at 1.0 atmospheric pressure by applying the Berendsen algorithm. The temperature and pressure coupling constants were set to 2.0 and 1.0 ps, respectively.
Additional information
How to cite this article: Huang, J. et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat. Commun. 3:911 doi: 10.1038/ncomms1919 (2012).
References
Das, S., Dixon, J. E. & Cho, W. Membrane-binding and activation mechanism of PTEN. Proc. Natl Acad. Sci. USA 100, 7491–7496 (2003).
Vazquez, F. et al. Tumor suppressor PTEN acts through dynamic interaction with the plasma membrane. Proc. Natl Acad. Sci. USA 103, 3633–3638 (2006).
Rahdar, M. et al. A phosphorylation-dependent intramolecular interaction regulates the membrane association and activity of the tumor suppressor PTEN. Proc. Natl Acad. Sci. USA 106, 480–485 (2009).
Lee, J. O. et al. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell 99, 323–334 (1999).
Georgescu, M. M. et al. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res. 60, 7033–7038 (2000).
Campbell, R. B., Liu, F. & Ross, A. H. Alloste1ric activation of PTEN phosphatase by phosphatidylinositol 4,5-bisphosphate. J. Biol. Chem. 278, 33617–33620 (2003).
Walker, S. M., Leslie, N. R., Perera, N. M., Batty, I. H. & Downes, C. P. The tumour-suppressor function of PTEN requires an N-terminal lipid-binding motif. Biochem. J. 379, 301–307 (2004).
Odriozola, L., Singh, G., Hoang, T. & Chan, A. M. Regulation of PTEN activity by its carboxyl-terminal autoinhibitory domain. J. Biol. Chem. 282, 23306–23015 (2007).
Vazquez, F., Ramaswamy, S., Nakamura, N. & Sellers, W. R. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell Biol. 20, 5010–5018 (2000).
Vazquez, F. et al. Phosphorylation of the PTEN tail acts as an inhibitory switch by preventing its recruitment into a protein complex. J. Biol. Chem. 276, 48627–48630 (2001).
Yu, J. et al. PTEN regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 28, 21–33 (2009).
Palomero, T. et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 13, 1203–1210 (2007).
Chappell, W. H. et al. Increased protein expression of the PTEN tumor suppressor in the presence of constitutively active Notch-1. Cell Cycle 4, 1389–1395 (2005).
Okumura, K. et al. PCAF modulates PTEN activity. J. Biol. Chem. 281, 26562–26568 (2006).
Wang, X. et al. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128, 129–139 (2007).
Trotman, L. C. et al. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128, 141–156 (2007).
Maddika, S. et al. WWP2 is an E3 ubiquitin ligase for PTEN. Nat. Cell Biol. 13, 728–33 (2011).
Lee, S. R. et al. Reversible inactivation of the tumor suppressor PTEN by H2O2. J. Biol. Chem. 277, 20336–20342 (2002).
Kwak, Y. D. et al. NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol. Neurodegener. 5, 49 (2010).
Li, Z. et al. Regulation of PTEN by Rho small GTPases. Nat. Cell Biol. 7, 399–404 (2005).
Al-Khouri, A. M., Ma, Y., Togo, S. H., Williams, S. & Mustelin, T. Cooperative phosphorylation of the tumor suppressor phosphatase and tensin homologue (PTEN) by casein kinases and glycogen synthase kinase 3beta. J. Biol. Chem. 280, 35195–35202 (2005).
Miller, S. J., Lou, D. Y., Seldin, D. C., Lane, W. S. & Neel, B. G. Direct identification of PTEN phosphorylation sites. FEBS Lett. 528, 145–153 (2002).
Torres, J. & Pulido, R. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J. Biol. Chem. 276, 993–998 (2001).
Waite, K. A. & Eng, C. BMP2 exposure results in decreased PTEN protein degradation and increased PTEN levels. Hum. Mol. Genet. 12, 679–684 (2003).
Shen, L. et al. SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat. Struct. Mol. Biol. 13, 1069–1077 (2006).
Xu, Z. & Au, S. W. Mapping residues of SUMO precursors essential in differential maturation by SUMO-specific protease, SENP1. Biochem. J. 386, 325–330 (2005).
Bayer, P. et al. Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275–286 (1998).
Eilebrecht, S., Smet-Nocca, C., Wieruszeski, J. M. & Benecke, A. SUMO-1 possesses DNA binding activity. BMC Res. Notes 3, 146 (2010).
Liu, J. L. et al. Nuclear PTEN-mediated growth suppression is independent of Akt down-regulation. Mol. Cell Biol. 25, 6211–6224 (2005).
Geiss-Friedlander, R. & Melchior, F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 (2007).
Sumitomo, M. et al. Synergy in tumor suppression by direct interaction of neutral endopeptidase with PTEN. Cancer Cell 5, 67–78 (2004).
Sobko, A., Ma, H. & Firtel, R. A. Regulated SUMOylation and ubiquitination of DdMEK1 is required for proper chemotaxis. Dev. Cell 2, 745–756 (2002).
Ross, A. H. & Gericke, A. Phosphorylation keeps PTEN phosphatase closed for business. Proc. Natl Acad. Sci. USA 106, 1297–1298 (2009).
Iijima, M., Huang, Y. E., Luo, H. R., Vazquez, F. & Devreotes, P. N. Novel mechanism of PTEN regulation by its phosphatidylinositol 4,5-bisphosphate binding motif is critical for chemotaxis. J. Biol. Chem. 279, 16606–16613 (2004).
Palm, D. et al. The norepinephrine-driven metastasis development of PC-3 human prostate cancer cells in BALB/c nude mice is inhibited by beta-blockers. Int. J. Cancer 118, 2744–2749 (2006).
Cheng, J., Kang, X., Zhang, S. & Yeh, E. T. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131, 584–595 (2007).
Gruber, P. et al. Barbituric acid derivative BAS 02104951 inhibits PKCepsilon, PKCeta, PKCepsilon/RACK2 interaction, Elk-1 phosphorylation in HeLa and PKCepsilon and eta translocation in PC3 cells following TPA-induction. J. Biochem. 149, 331–336 (2011).
van Loo, G. et al. Caspases are not localized in mitochondria during life or death. Cell Death Differ. 9, 1207–1211 (2002).
Janetopoulos, C., Borleis, J., Vazquez, F., Iijima, M. & Devreotes, P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev. Cell 8, 467–477 (2005).
Ulrich, F. et al. Wnt11 functions in gastrulation by controlling cell cohesion through Rab5c and E-cadherin. Dev. Cell 9, 555–564 (2005).
Wang, J., Cieplak, P. & Kollman, P. A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 21, 1049–1074 (2000).
Miyamoto, S. & Kollman, P. A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13, 952–962 (1992).
Acknowledgements
This work was supported by grants National Key Program (973) for Basic Research of China (2011CB811303, 2012CB917101, 2009CB918404); National Natural Science Foundation (31071230, 91129726, 81171941); Shanghai Pujiang Program (10PJ1406600); Shanghai Talent Development Fund to Dr. JY(u); and Shanghai Committee of Science and Technology (11DZ2260200). We also acknowledge partial support from State Key Laboratory of Neuroscience, Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
J. H. performed most of the experiments, analysed data. J. Yan generated PTEN lentivirus and infected cells, did some staining of cells and tissues. J. Z. and T. S. built up the three-dimensional structural model of SUMO1–PTEN and provided Figs 4a and 6a. Y. W. and S. Z. performed animal experiments. C. Z., C. C. and X. L. helped with all experiments. J. C. made SENP1−/− MEFs. G. C., T. M. and G. F. analysed data. J. Yu designed and supervised all aspects of this work and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1-S9 and Supplementary Methods (PDF 3624 kb)
Rights and permissions
About this article
Cite this article
Huang, J., Yan, J., Zhang, J. et al. SUMO1 modification of PTEN regulates tumorigenesis by controlling its association with the plasma membrane. Nat Commun 3, 911 (2012). https://doi.org/10.1038/ncomms1919
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms1919
This article is cited by
-
N4-acetylcytidine modifies primary microRNAs for processing in cancer cells
Cellular and Molecular Life Sciences (2024)
-
Glucose-mediated N-glycosylation of RPTPα affects its subcellular localization and Src activation
Oncogene (2023)
-
A DNA copy number alteration classifier as a prognostic tool for prostate cancer patients
British Journal of Cancer (2023)
-
SUMOylation of RALY promotes vasculogenic mimicry in glioma cells via the FOXD1/DKK1 pathway
Cell Biology and Toxicology (2023)
-
TRIM25 activates AKT/mTOR by inhibiting PTEN via K63-linked polyubiquitination in non-small cell lung cancer
Acta Pharmacologica Sinica (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.