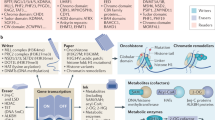
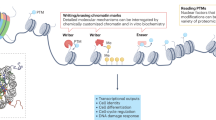
Abstract
Chromatin is extensively chemically modified and thereby acts as a dynamic signaling platform controlling gene function. Chromatin regulation is integral to cell differentiation, lineage commitment and organism development, whereas chromatin dysregulation can lead to age-related and neurodegenerative disorders as well as cancer. Investigating chromatin biology presents a unique challenge, as the issue spans many disciplines, including cell and systems biology, biochemistry and molecular biophysics. In recent years, the application of chemical biology methods for investigating chromatin processes has gained considerable traction. Indeed, chemical biologists now have at their disposal powerful chemical tools that allow chromatin biology to be scrutinized at the level of the cell all the way down to the single chromatin fiber. Here we present recent examples of how this rapidly expanding palette of chemical tools is being used to paint a detailed picture of chromatin function in organism development and disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Woodcock, C.L. & Ghosh, R.P. Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2, a000596 (2010).
Jenuwein, T. & Allis, C.D. Translating the histone code. Science 293, 1074–1080 (2001).
Chi, P., Allis, C.D. & Wang, G.G. Covalent histone modifications–miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 10, 457–469 (2010).
Cole, P.A. Chemical probes for histone-modifying enzymes. Nat. Chem. Biol. 4, 590–597 (2008).
Chatterjee, C. & Muir, T.W. Chemical approaches for studying histone modifications. J. Biol. Chem. 285, 11045–11050 (2010).
Filion, G.J. et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224 (2010).
Ernst, J. & Kellis, M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 (2010).
Kharchenko, P.V. et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471, 480–485 (2011).
Taverna, S.D., Li, H., Ruthenburg, A.J., Allis, C.D. & Patel, D.J. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Taunton, J., Hassig, C.A. & Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 (1996).
Mund, C. & Lyko, F. Epigenetic cancer therapy: Proof of concept and remaining challenges. Bioessays 32, 949–957 (2010).
Shahbazian, M.D. & Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76, 75–100 (2007).
Tóth, K.F. et al. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 117, 4277–4287 (2004).
Llères, D., James, J., Swift, S., Norman, D.G. & Lamond, A.I. Quantitative analysis of chromatin compaction in living cells using FLIM-FRET. J. Cell Biol. 187, 481–496 (2009).
Marks, P.A. & Breslow, R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat. Biotechnol. 25, 84–90 (2007).
Huangfu, D. et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 (2008).
Sternson, S.M., Wong, J.C., Grozinger, C.M. & Schreiber, S.L. Synthesis of 7200 small molecules based on a substructural analysis of the histone deacetylase inhibitors trichostatin and trapoxin. Org. Lett. 3, 4239–4242 (2001).
Bieliauskas, A.V. & Pflum, M.K.H. Isoform-selective histone deacetylase inhibitors. Chem. Soc. Rev. 37, 1402–1413 (2008).
Bradner, J.E. et al. Chemical phylogenetics of histone deacetylases. Nat. Chem. Biol. 6, 238–243 (2010).
Bantscheff, M. et al. Chemoproteomics profiling of HDAC inhibitors reveals selective targeting of HDAC complexes. Nat. Biotechnol. 29, 255–265 (2011).
Greiner, D., Bonaldi, T., Eskeland, R., Roemer, E. & Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat. Chem. Biol. 1, 143–145 (2005).
McGarvey, K.M. et al. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 66, 3541–3549 (2006).
Kubicek, S. et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007).
Chang, Y. et al. Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294. Nat. Struct. Mol. Biol. 16, 312–317 (2009).
Vedadi, M. et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat. Chem. Biol. 7, 566–574 (2011).
Shi, Y. et al. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell 2, 525–528 (2008).
Min, J., Feng, Q., Li, Z.Z., Zhang, Y. & Xu, R.M. Structure of the catalytic domain of human Dot1L, a Non-SET domain nucleosomal histone methyltransferase. Cell 112, 711–723 (2003).
Feng, Q. et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 12, 1052–1058 (2002).
Ng, H.H. et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 16, 1518–1527 (2002).
van Leeuwen, F., Gafken, P.R. & Gottschling, D.E. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109, 745–756 (2002).
Okada, Y. et al. hDOT1L links histone methylation to leukemogenesis. Cell 121, 167–178 (2005).
Krivtsov, A.V. et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell 14, 355–368 (2008).
Daigle, S.R. et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65 (2011).
Bishop, A.C. et al. Design of allele-specific inhibitors to probe protein kinase signaling. Curr. Biol. 8, 257–266 (1998).
Lin, Q., Jiang, F.Y., Schultz, P.G. & Gray, N.S. Design of allele-specific protein methyltransferase inhibitors. J. Am. Chem. Soc. 123, 11608–11613 (2001).
Suganuma, T. & Workman, J.L. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80, 473–499 (2011).
Moazed, D. Mechanisms for the inheritance of chromatin states. Cell 146, 510–518 (2011).
Kim, J. et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 7, 397–403 (2006).
Fischle, W., Franz, H., Jacobs, S.A., Allis, C.D. & Khorasanizadeh, S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J. Biol. Chem. 283, 19626–19635 (2008).
Fischle, W. et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17, 1870–1881 (2003).
Fischle, W. et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122 (2005).
Flanagan, J.F. et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature 438, 1181–1185 (2005).
Ramón-Maiques, S. et al. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc. Natl. Acad. Sci. USA 104, 18993–18998 (2007).
Garske, A.L., Craciun, G. & Denu, J.M. A combinatorial H4 tail library for exploring the histone code. Biochemistry 47, 8094–8102 (2008).
Garske, A.L. et al. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat. Chem. Biol. 6, 283–290 (2010).
Ruthenburg, A.J., Li, H., Patel, D.J. & Allis, C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 8, 983–994 (2007).
Jacobson, R.H., Ladurner, A.G., King, D.S. & Tjian, R. Structure and function of a human TAFII250 double bromodomain module. Science 288, 1422–1425 (2000).
Ruthenburg, A.J. et al. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 145, 692–706 (2011).
Schulze, W.X. & Mann, M. A novel proteomic screen for peptide-protein interactions. J. Biol. Chem. 279, 10756–10764 (2004).
Vermeulen, M. et al. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131, 58–69 (2007).
Vermeulen, M. et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142, 967–980 (2010).
Bartke, T. et al. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143, 470–484 (2010).
Nikolov, M. et al. Chromatin affinity purification and quantitative mass spectrometry defining the interactome of histone modification patterns. Mol. Cell. Proteomics 10 (2011).
Shi, X. et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 442, 96–99 (2006).
Li, B. et al. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science 316, 1050–1054 (2007).
Zeng, L. et al. Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b. Nature 466, 258–262 (2010).
Wells, J.A. & McClendon, C.L. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature 450, 1001–1009 (2007).
Zeng, L. et al. Selective small molecules blocking HIV-1 Tat and coactivator PCAF association. J. Am. Chem. Soc. 127, 2376–2377 (2005).
Sachchidanand et al. Target structure-based discovery of small molecules that block human p53 and CREB binding protein association. Chem. Biol. 13, 81–90 (2006).
Borah, J.C. et al. A small molecule binding to the coactivator CREB-binding protein blocks apoptosis in cardiomyocytes. Chem. Biol. 18, 531–541 (2011).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Nicodeme, E. et al. Suppression of inflammation by a synthetic histone mimic. Nature 468, 1119–1123 (2010).
Morinière, J. et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 461, 664–668 (2009).
Miyoshi, S., Ooike, S., Iwata, K., Hikawa, H. & Sugaraha, K. Antitumor agent. International patent PCT/JP2008/073864 (WO/2009/084693) (2009).
Reynoird, N. et al. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J. 29, 2943–2952 (2010).
Hargreaves, D.C., Horng, T. & Medzhitov, R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell 138, 129–145 (2009).
Herold, J.M. et al. Small-molecule ligands of methyl-lysine binding proteins. J. Med. Chem. 54, 2504–2511 (2011).
Li, S. & Shogren-Knaak, M.A. Cross-talk between histone H3 tails produces cooperative nucleosome acetylation. Proc. Natl. Acad. Sci. USA 105, 18243–18248 (2008).
Li, S. & Shogren-Knaak, M.A. The Gcn5 bromodomain of the SAGA complex facilitates cooperative and cross-tail acetylation of nucleosomes. J. Biol. Chem. 284, 9411–9417 (2009).
Margueron, R. et al. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461, 762–767 (2009).
Li, B. et al. Histone H3 lysine 36 dimethylation (H3K36me2) is sufficient to recruit the Rpd3s histone deacetylase complex and to repress spurious transcription. J. Biol. Chem. 284, 7970–7976 (2009).
Ng, H.H., Xu, R.M., Zhang, Y. & Struhl, K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J. Biol. Chem. 277, 34655–34657 (2002).
McGinty, R.K., Kim, J., Chatterjee, C., Roeder, R.G. & Muir, T.W. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812–816 (2008).
McGinty, R.K. et al. Structure activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS Chem. Biol. 4, 958–968 (2009).
Chatterjee, C., McGinty, R.K., Fierz, B. & Muir, T.W. Disulfide directed histone ubiquitylation reveals plasticity in hDot1L stimulation. Nat. Chem. Biol. 6, 267–269 (2010).
Kim, J. et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137, 459–471 (2009).
Sun, Z.W. & Allis, C.D. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418, 104–108 (2002).
Canzio, D. et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell 41, 67–81 (2011).
Shogren-Knaak, M. et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Lu, X. et al. The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat. Struct. Mol. Biol. 15, 1122–1124 (2008).
Fierz, B. et al. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat. Chem. Biol. 7, 113–119 (2011).
Polach, K.J. & Widom, J. Mechanism of protein access to specific DNA-sequences in chromatin - a dynamic equilibrium-model for gene-regulation. J. Mol. Biol. 254, 130–149 (1995).
Li, G. & Widom, J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 11, 763–769 (2004).
Gansen, A. et al. Nucleosome disassembly intermediates characterized by single-molecule FRET. Proc. Natl. Acad. Sci. USA 106, 15308–15313 (2009).
Hall, M.A. et al. High-resolution dynamic mapping of histone-DNA interactions in a nucleosome. Nat. Struct. Mol. Biol. 16, 124–129 (2009).
Luger, K., Mader, A.W., Richmond, R.K., Sargent, D.F. & Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Neumann, H. et al. A method for genetically installing site-specific acetylation in recombinant histones defines the effects of H3 K56 acetylation. Mol. Cell 36, 153–163 (2009).
Simon, M. et al. Histone fold modifications control nucleosome unwrapping and disassembly. Proc. Natl. Acad. Sci. USA 108, 12711–12716 (2011).
Dawson, P.E., Muir, T.W., Clark-Lewis, I. & Kent, S.B.H. Synthesis of proteins by native chemical ligation. Science 266, 776–779 (1994).
Muir, T.W., Sondhi, D. & Cole, P.A. Expressed protein ligation: a general method for protein engineering. Proc. Natl. Acad. Sci. USA 95, 6705–6710 (1998).
Kee, J.M., Villani, B., Carpenter, L.R. & Muir, T.W. Development of stable phosphohistidine analogues. J. Am. Chem. Soc. 132, 14327–14329 (2010).
Moyle, P.M. & Muir, T.W. Method for the synthesis of mono-ADP-ribose conjugated peptides. J. Am. Chem. Soc. 132, 15878–15880 (2010).
Chatterjee, C., McGinty, R.K., Pellois, J.P. & Muir, T.W. Auxiliary-mediated site-specific peptide ubiquitylation. Angew. Chem. Int. Edn Engl. 46, 2814–2818 (2007).
Ajish Kumar, K.S., Haj-Yahya, M., Olschewski, D., Lashuel, H.A. & Brik, A. Highly efficient and chemoselective peptide ubiquitylation. Angew. Chem. Int. Edn Engl. 48, 8090–8094 (2009).
Simon, M.D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
Li, F. et al. A direct method for site-specific protein acetylation. Angew. Chem. Int. Edn Engl. 50, 9611–9614 (2011).
Neumann, H., Peak-Chew, S.Y. & Chin, J.W. Genetically encoding N-epsilon-acetyllysine in recombinant proteins. Nat. Chem. Biol. 4, 232–234 (2008).
Ito, T., Bulger, M., Pazin, M.J., Kobayashi, R. & Kadonaga, J.T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 (1997).
Oudet, P., Gross-Bellard, M. & Chambon, P. Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4, 281–300 (1975).
Zheng, C. & Hayes, J.J. Intra- and inter-nucleosomal protein-DNA interactions of the core histone tail domains in a model system. J. Biol. Chem. 278, 24217–24224 (2003).
Acknowledgements
Some of the work discussed in this article was performed in the authors' group and was supported by the US National Institutes of Health and by the Swiss National Science Foundation, No. PA00P3-129130 (B.F.). We thank members of the Muir laboratory for critical reading of this article and apologize to the researchers whose work could not be cited because of space restraints.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Fierz, B., Muir, T. Chromatin as an expansive canvas for chemical biology. Nat Chem Biol 8, 417–427 (2012). https://doi.org/10.1038/nchembio.938
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.938
This article is cited by
-
Reconstitution of Drosophila and human chromatins by wheat germ cell-free co-expression system
BMC Biotechnology (2020)
-
Mechanism of biomolecular recognition of trimethyllysine by the fluorinated aromatic cage of KDM5A PHD3 finger
Communications Chemistry (2020)
-
Histone H3.3 phosphorylation amplifies stimulation-induced transcription
Nature (2020)
-
Acetylation of the histone H3 tail domain regulates base excision repair on higher-order chromatin structures
Scientific Reports (2019)
-
The nucleophilic amino group of lysine is central for histone lysine methyltransferase catalysis
Communications Chemistry (2019)