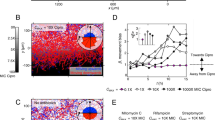
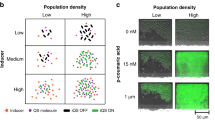
Abstract
Here we show that bacterial communication through indole signaling induces persistence, a phenomenon in which a subset of an isogenic bacterial population tolerates antibiotic treatment. We monitor indole-induced persister formation using microfluidics and identify the role of oxidative-stress and phage-shock pathways in this phenomenon. We propose a model in which indole signaling 'inoculates' a bacterial subpopulation against antibiotics by activating stress responses, leading to persister formation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Balaban, N.Q. et al. Science 305, 1622–1625 (2004).
Lewis, K. Nat. Rev. Microbiol. 5, 48–56 (2007).
Smith, P.A. & Romesberg, F.E. Nat. Chem. Biol. 3, 549–556 (2007).
Allison, K.R., Brynildsen, M.P. & Collins, J.J. Nature 473, 216–220 (2011).
Levin, B.R. & Rozen, D.E. Nat. Rev. Microbiol. 4, 556–562 (2006).
Gefen, O. & Balaban, N.Q. FEMS Microbiol. Rev. 33, 704–717 (2009).
Keren, I. et al. FEMS Microbiol. Lett. 230, 13–18 (2004).
Moyed, H.S. & Bertrand, K.P. J. Bacteriol. 155, 768–775 (1983).
Wang, X. & Wood, T.K. Appl. Environ. Microbiol. 77, 5577–5583 (2011).
Wang, X. et al. Nat. Chem. Biol. 7, 359–366 (2011).
Bassler, B.L. & Losick, R. Cell 125, 237–246 (2006).
Wang, D., Ding, X.D. & Rather, P.N. J. Bacteriol. 183, 4210–4216 (2001).
Martino, P.D. et al. Can. J. Microbiol. 49, 443–449 (2003).
Lee, J. et al. ISME J. 2, 1007–1023 (2008).
Han, T.H. et al. Res. Microbiol. 162, 108–116 (2010).
Yanofsky, C., Horn, V. & Gollnick, P. J. Bacteriol. 173, 6009–6017 (1991).
Piñero-Fernandez, S., Chimerel, C., Keyser, U.F. & Summers, D.K. J. Bacteriol. 193, 1793–1798 (2011).
Garbe, T.R., Kobayashi, M. & Yukawa, H. Arch. Microbiol. 173, 78–82 (2000).
Hirakawa, H. et al. Mol. Microbiol. 55, 1113–1126 (2005).
Lee, H.H., Molla, M.N., Cantor, C.R. & Collins, J.J. Nature 467, 82–85 (2010).
Weiner, L. & Model, P. Proc. Natl. Acad. Sci. USA 91, 2191–2195 (1994).
Dukan, S. & Nystrom, T. J. Biol. Chem. 274, 26027–26032 (1999).
Storz, G., Tartaglia, L.A. & Ames, B.N. Antonie Van Leeuwenhoek 58, 157–161 (1990).
Crawford, D.R. & Davies, K.J. Environ. Health Perspect. 102 (suppl.), 25–28 (1994).
Rotem, E. et al. Proc. Natl. Acad. Sci. USA 107, 12541–12546 (2010).
Acknowledgements
We would like to thank R.H.W. Lam for help with microfluidics. This work was supported by funding from the US National Science Foundation, the US National Institutes of Health Director's Pioneer Award Program and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
N.M.V., K.R.A., A.S.K. and J.J.C. designed experiments, discussed results and contributed to the manuscript. N.M.V. performed all experiments. N.M.V., K.R.A. and A.S.K. analyzed data. A.S.K. developed the microfluidics platform and performed the microfluidic experiments.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results and Supplementary Note 1 (PDF 1456 kb)
Supplementary Movie 1
Vega_SuppMovie1 (MOV 53270 kb)
Supplementary Movie 2
Vega_SuppMovie2 (MOV 42497 kb)
Supplementary Movie 3
Vega_SuppMovie3 (MOV 53170 kb)
Rights and permissions
About this article
Cite this article
Vega, N., Allison, K., Khalil, A. et al. Signaling-mediated bacterial persister formation. Nat Chem Biol 8, 431–433 (2012). https://doi.org/10.1038/nchembio.915
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.915