Abstract
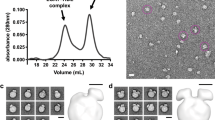
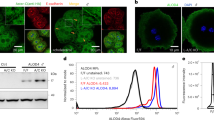
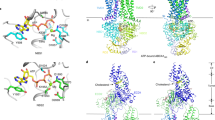
Human cholesteryl ester transfer protein (CETP) mediates the net transfer of cholesteryl ester mass from atheroprotective high-density lipoproteins to atherogenic low-density lipoproteins by an unknown mechanism. Delineating this mechanism would be an important step toward the rational design of new CETP inhibitors for treating cardiovascular diseases. Using EM, single-particle image processing and molecular dynamics simulation, we discovered that CETP bridges a ternary complex with its N-terminal β-barrel domain penetrating into high-density lipoproteins and its C-terminal domain interacting with low-density lipoprotein or very-low-density lipoprotein. In our mechanistic model, the CETP lipoprotein-interacting regions, which are highly mobile, form pores that connect to a hydrophobic central cavity, thereby forming a tunnel for transfer of neutral lipids from donor to acceptor lipoproteins. These new insights into CETP transfer provide a molecular basis for analyzing mechanisms for CETP inhibition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Barter, P.J. et al. Cholesteryl ester transfer protein: a novel target for raising HDL and inhibiting atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 23, 160–167 (2003).
Camejo, G., Waich, S., Quintero, G., Berrizbeitia, M.L. & Lalaguna, F. The affinity of low density lipoproteins for an arterial macromolecular complex. A study in ischemic heart disease and controls. Atherosclerosis 24, 341–354 (1976).
Gordon, T., Castelli, W.P., Hjortland, M.C., Kannel, W.B. & Dawber, T.R. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am. J. Med. 62, 707–714 (1977).
Hayek, T. et al. Hypertriglyceridemia and cholesteryl ester transfer protein interact to dramatically alter high density lipoprotein levels, particle sizes, and metabolism. Studies in transgenic mice. J. Clin. Invest. 92, 1143–1152 (1993).
Brown, M.L. et al. Molecular basis of lipid transfer protein deficiency in a family with increased high-density lipoproteins. Nature 342, 448–451 (1989).
Inazu, A. et al. Increased high-density lipoprotein levels caused by a common cholesteryl-ester transfer protein gene mutation. N. Engl. J. Med. 323, 1234–1238 (1990).
Niesor, E.J. Different effects of compounds decreasing cholesteryl ester transfer protein activity on lipoprotein metabolism. Curr. Opin. Lipidol. 22, 288–295 (2011).
Miyares, M.A. Anacetrapib and dalcetrapib: two novel cholesteryl ester transfer protein inhibitors. Ann. Pharmacother. 45, 84–94 (2011).
Kappelle, P.J., van Tol, A., Wolffenbuttel, B.H. & Dullaart, R.P. Cholesteryl ester transfer protein inhibition in cardiovascular risk management: ongoing trials will end the confusion. Cardiovasc. Ther. 29, e89–e99 (2010).
Qiu, X. et al. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat. Struct. Mol. Biol. 14, 106–113 (2007).
Barter, P.J. & Jones, M.E. Kinetic studies of the transfer of esterified cholesterol between human plasma low and high density lipoproteins. J. Lipid Res. 21, 238–249 (1980).
Ihm, J., Quinn, D.M., Busch, S.J., Chataing, B. & Harmony, J.A. Kinetics of plasma protein-catalyzed exchange of phosphatidylcholine and cholesteryl ester between plasma lipoproteins. J. Lipid Res. 23, 1328–1341 (1982).
Tall, A.R. Plasma cholesteryl ester transfer protein. J. Lipid Res. 34, 1255–1274 (1993).
Zhang, L. et al. Morphology and structure of lipoproteins revealed by an optimized negative-staining protocol of electron microscopy. J. Lipid Res. 52, 175–184 (2011).
Chen, B. et al. Apolipoprotein AI tertiary structures determine stability and phospholipid-binding activity of discoidal high-density lipoprotein particles of different sizes. Protein Sci. 18, 921–935 (2009).
Silva, R.A. et al. Structure of apolipoprotein A-I in spherical high density lipoproteins of different sizes. Proc. Natl. Acad. Sci. USA 105, 12176–12181 (2008).
Zhang, L. et al. An optimized negative-staining protocol of electron microscopy for apoE4·POPC lipoprotein. J. Lipid Res. 51, 1228–1236 (2010).
Ohi, M., Li, Y., Cheng, Y. & Walz, T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol. Proced. Online 6, 23–34 (2004).
Ren, G., Reddy, V.S., Cheng, A., Melnyk, P. & Mitra, A.K. Visualization of a water-selective pore by electron crystallography in vitreous ice. Proc. Natl. Acad. Sci. USA 98, 1398–1403 (2001).
Ren, G., Cheng, A., Reddy, V., Melnyk, P. & Mitra, A.K. Three-dimensional fold of the human AQP1 water channel determined at 4 Å resolution by electron crystallography of two-dimensional crystals embedded in ice. J. Mol. Biol. 301, 369–387 (2000).
Ren, G. et al. Model of human low-density lipoprotein and bound receptor based on CryoEM. Proc. Natl. Acad. Sci. USA 107, 1059–1064 (2010).
Adrian, M., Dubochet, J., Fuller, S.D. & Harris, J.R. Cryo-negative staining. Micron 29, 145–160 (1998).
Hayat, M.A. & Miller, S.E. Negative Staining (McGraw-Hill, 1990).
Morton, R.E. & Greene, D.J. The surface cholesteryl ester content of donor and acceptor particles regulates CETP: a liposome-based approach to assess the substrate properties of lipoproteins. J. Lipid Res. 44, 1364–1372 (2003).
Swenson, T.L., Brocia, R.W. & Tall, A.R. Plasma cholesteryl ester transfer protein has binding sites for neutral lipids and phospholipids. J. Biol. Chem. 263, 5150–5157 (1988).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Liu, J., Bartesaghi, A., Borgnia, M.J., Sapiro, G. & Subramaniam, S. Molecular architecture of native HIV-1 gp120 trimers. Nature 455, 109–113 (2008).
Flemming, D., Thierbach, K., Stelter, P., Böttcher, B. & Hurt, E. Precise mapping of subunits in multiprotein complexes by a versatile electron microscopy label. Nat. Struct. Mol. Biol. 17, 775–778 (2010).
Phillips, J.C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005).
Atilgan, A.R. et al. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 80, 505–515 (2001).
Doruker, P., Atilgan, A.R. & Bahar, I. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: application to α-amylase inhibitor. Proteins 40, 512–524 (2000).
Weers, P.M. & Ryan, R.O. Apolipophorin III: role model apolipoprotein. Insect Biochem. Mol. Biol. 36, 231–240 (2006).
Saito, H. et al. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J. Biol. Chem. 278, 23227–23232 (2003).
Mehta, R., Gantz, D.L. & Gursky, O. Human plasma high-density lipoproteins are stabilized by kinetic factors. J. Mol. Biol. 328, 183–192 (2003).
Han, M. et al. Disruption of human plasma high-density lipoproteins by streptococcal serum opacity factor requires labile apolipoprotein A-I. Biochemistry 48, 1481–1487 (2009).
Wang, S., Kussie, P., Deng, L. & Tall, A. Defective binding of neutral lipids by a carboxyl-terminal deletion mutant of cholesteryl ester transfer protein. Evidence for a carboxyl-terminal cholesteryl ester binding site essential for neutral lipid transfer activity. J. Biol. Chem. 270, 612–618 (1995).
Grigorieff, N. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157, 117–125 (2007).
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Galkin, V.E., Orlova, A., Cherepanova, O., Lebart, M.C. & Egelman, E.H. High-resolution cryo-EM structure of the F-actin-fimbrin/plastin ABD2 complex. Proc. Natl. Acad. Sci. USA 105, 1494–1498 (2008).
Böttcher, B., Wynne, S.A. & Crowther, R.A. Determination of the fold of the core protein of hepatitis B virus by electron cryomicroscopy. Nature 386, 88–91 (1997).
Acknowledgements
We thank D.A. Agard, I. Bahar and K. Dill for valuable discussions and A. Cheng and J. Song for helpful comments. This work was supported by Basic Energy Sciences–US Department of Energy (DE-AC02-05CH11231) and the W. M. Keck Foundation (no. 011808); the Keygrant Project of the Chinese Ministry of Education no. 708082 (S.Z.); US National Institutes of Health grant NIH-HL077268 and Tobacco-Related Disease Research Program of California grant 16FT-0163 (M.O.).
Author information
Authors and Affiliations
Contributions
G.R., K.H.W. and X.Q. initiated and designed the project; X.Q. provided the soluble CETP; H.J.P. and R.M.K. contributed the LDL and VLDL; K.-A.R., M.O. and G.C. provided HDL; L.Z., F.Y. and G.R. collected the data; S.Z., D.L. and G.R. conducted the molecular simulation; G.R. solved the structure; G.R., L.Z., M.A.C. and X.Q. analyzed and interpreted the data; G.R. and S.Z. drafted the initial manuscript; M.A.C., H.J.P., K.H.W., K.-A.R., R.M.K., X.Q. and L.Z. discussed and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 17930 kb)
Rights and permissions
About this article
Cite this article
Zhang, L., Yan, F., Zhang, S. et al. Structural basis of transfer between lipoproteins by cholesteryl ester transfer protein. Nat Chem Biol 8, 342–349 (2012). https://doi.org/10.1038/nchembio.796
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.796
This article is cited by
-
Structure-based mechanism and inhibition of cholesteryl ester transfer protein
Current Atherosclerosis Reports (2023)
-
LoTToR: An Algorithm for Missing-Wedge Correction of the Low-Tilt Tomographic 3D Reconstruction of a Single-Molecule Structure
Scientific Reports (2020)
-
Single-Molecule 3D Images of “Hole-Hole” IgG1 Homodimers by Individual-Particle Electron Tomography
Scientific Reports (2019)
-
Lipid transfer proteins: the lipid commute via shuttles, bridges and tubes
Nature Reviews Molecular Cell Biology (2019)
-
An Algorithm for Enhancing the Image Contrast of Electron Tomography
Scientific Reports (2018)