Abstract
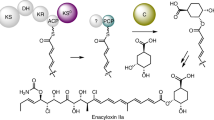
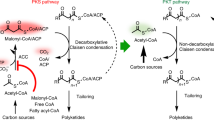
Polyketides are structurally diverse and medically important natural products that have various biological activities. During biosynthesis, chain elongation uses activated dicarboxylic acid building blocks, and their availability therefore limits side chain variation in polyketides. Recently, the crotonyl-CoA carboxylase-reductase (CCR) class of enzymes was identified in primary metabolism and was found to be involved in extender-unit biosynthesis of polyketides. These enzymes are, in theory, capable of forming dicarboxylic acids that show any side chain from the respective unsaturated fatty acid precursor. To our knowledge, we here report the first crystal structure of a CCR, the hexylmalonyl-CoA synthase from Streptomyces sp. JS360, in complex with its substrate. Structural analysis and biochemical characterization of the enzyme, including active site mutations, are reported. Our analysis reveals how primary metabolic CCRs can evolve to produce various dicarboxylic acid building blocks, setting the stage to use CCRs for the production of unique extender units and, consequently, altered polyketides.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hertweck, C. The biosynthetic logic of polyketide diversity. Angew. Chem. Int. Edn Engl. 48, 4688–4716 (2009).
Newman, D.J. & Cragg, G.M. Natural products as sources of new drugs over the last 25 years. J. Nat. Prod. 70, 461–477 (2007).
Staunton, J. & Weissman, K.J. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 18, 380–416 (2001).
Wenzel, S.C. et al. On the biosynthetic origin of methoxymalonyl-acyl carrier protein, the substrate for incorporation of “glycolate” units into ansamitocin and soraphen A. J. Am. Chem. Soc. 128, 14325–14336 (2006).
Dorrestein, P.C. et al. The bifunctional glyceryl transferase/phosphatase OzmB belonging to the HAD superfamily that diverts 1,3-bisphosphoglycerate into polyketide biosynthesis. J. Am. Chem. Soc. 128, 10386–10387 (2006).
Chan, Y.A. et al. Hydroxymalonyl-acyl carrier protein (ACP) and aminomalonyl-ACP are two additional type I polyketide synthase extender units. Proc. Natl. Acad. Sci. USA 103, 14349–14354 (2006).
Erb, T.J. et al. Synthesis of C5-dicarboxylic acids from C2-units involving crotonyl-CoA carboxylase/reductase: the ethylmalonyl-CoA pathway. Proc. Natl. Acad. Sci. USA 104, 10631–10636 (2007).
Erb, T.J., Brecht, V., Fuchs, G., Muller, M. & Alber, B.E. Carboxylation mechanism and stereochemistry of crotonyl-CoA carboxylase/reductase, a carboxylating enoyl-thioester reductase. Proc. Natl. Acad. Sci. USA 106, 8871–8876 (2009).
Rachid, S. et al. Mining the cinnabaramide biosynthetic pathway to generate novel proteasome inhibitors. ChemBioChem 12, 922–931 (2011).
Buntin, K. et al. Biosynthesis of thuggacins in myxobacteria: comparative cluster analysis reveals basis for natural product structural diversity. Chem. Biol. 17, 342–356 (2010).
Eustáquio, A.S. et al. Biosynthesis of the salinosporamide A polyketide synthase substrate chloroethylmalonyl-coenzyme A from S-adenosyl-L-methionine. Proc. Natl. Acad. Sci. USA 106, 12295–12300 (2009).
Mo, S. et al. Biosynthesis of the allylmalonyl-CoA extender unit for the FK506 polyketide synthase proceeds through a dedicated polyketide synthase and facilitates the mutasynthesis of analogues. J. Am. Chem. Soc. 133, 976–985 (2011).
Goranovic, D. et al. Origin of the allyl group in FK506 biosynthesis. J. Biol. Chem. 285, 14292–14300 (2010).
Qu, X. et al. Cloning, sequencing and characterization of the biosynthetic gene cluster of sanglifehrin A, a potent cyclophilin inhibitor. Mol. Biosyst. 7, 852–861 (2011).
Kopp, M. et al. Insights into the complex biosynthesis of the leupyrrins in Sorangium cellulosum So ce690. Mol. Biosyst. 7, 1549–1563 (2011).
Song, L. et al. Type III polyketide synthase I2-ketoacyl-ACP starter unit and ethylmalonyl-CoA extender unit selectivity discovered by Streptomyces coelicolor genome mining. J. Am. Chem. Soc. 128, 14754–14755 (2006).
Xu, Z., Ding, L. & Hertweck, C. A branched extender unit shared between two orthogonal polyketide pathways in an endophyte. Angew. Chem. Int. Edn Engl. 50, 4667–4670 (2011).
Wilson, M.C. et al. Structure and biosynthesis of the marine Streptomycete ansamycin ansalactam A and its distinctive branched chain polyketide extender unit. J. Am. Chem. Soc. published online, doi:10.1021/ja109226s (19 January 2011).
Yoo, H.G. et al. Characterization of 2-octenoyl-CoA carboxylase/reductase utilizing pteB from Streptomyces avermitilis. Biosci. Biotechnol. Biochem. 75, 1191–1193 (2011).
Xu, L.H. et al. Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis. J. Biol. Chem. 285, 16844–16853 (2010).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Verdonk, M.L. et al. Modeling water molecules in protein-ligand docking using GOLD. J. Med. Chem. 48, 6504–6515 (2005).
Li, X., Jayachandran, S., Nguyen, H.H. & Chan, M.K. Structure of the Nitrosomonas europaea Rh protein. Proc. Natl. Acad. Sci. USA 104, 19279–19284 (2007).
Esposito, L. et al. Crystal structure of a ternary complex of the alcohol dehydrogenase from Sulfolobus solfataricus. Biochemistry 42, 14397–14407 (2003).
Porté, S. et al. Three-dimensional structure and enzymatic function of proapoptotic human p53-inducible quinone oxidoreductase PIG3. J. Biol. Chem. 284, 17194–17205 (2009).
Weissman, K.J. & Leadlay, P.F. Combinatorial biosynthesis of reduced polyketides. Nat. Rev. Microbiol. 3, 925–936 (2005).
Acknowledgements
D.W.H. acknowledges support from the Fonds der Chemischen Industrie. Research in R.M.'s laboratory was funded by the Deutsche Forschungsgemeinschaft and the Bundesministerium für Bildung und Forschung (BMBF). We thank V. Wray for critically reviewing this manuscript. We also would like to thank T. Hoffmann for technical support, K. Harmrolfs for the NMR measurement and S.C. Wenzel for advice during the course of this project. In addition, we appreciate the work of A. Ullrich and K. Schultz from U. Kazmaier's work group for the syntheses of 2-octenoyl-CoA and 2-octenoyl-SNAC.
Author information
Authors and Affiliations
Contributions
N.Q. performed protein crystallization, crystallographic data collection and structure determination. L.H. performed gene cloning, protein heterologous expression and purification experiments as well as in vitro characterization of proteins and generation of corresponding point mutations. S.R. contributed initial advice regarding protein expression. R.M. and D.W.H. designed the study, and R.M. wrote the manuscript together with N.Q., L.H. and D.W.H. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Results (PDF 1683 kb)
Rights and permissions
About this article
Cite this article
Quade, N., Huo, L., Rachid, S. et al. Unusual carbon fixation gives rise to diverse polyketide extender units. Nat Chem Biol 8, 117–124 (2012). https://doi.org/10.1038/nchembio.734
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.734
This article is cited by
-
The multicatalytic compartment of propionyl-CoA synthase sequesters a toxic metabolite
Nature Chemical Biology (2018)
-
A conserved threonine prevents self-intoxication of enoyl-thioester reductases
Nature Chemical Biology (2017)
-
Warhead biosynthesis and the origin of structural diversity in hydroxamate metalloproteinase inhibitors
Nature Communications (2017)
-
Characterization and engineering of the biosynthesis gene cluster for antitumor macrolides PM100117 and PM100118 from a marine actinobacteria: generation of a novel improved derivative
Microbial Cell Factories (2016)
-
Effect of cerulenin on fatty acid composition and gene expression pattern of DHA-producing strain Colwellia psychrerythraea strain 34H
Microbial Cell Factories (2016)