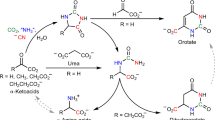
Abstract
Although amino acids are known precursors of purines, a pathway for the direct recycling of amino acids from purines has never been described at the molecular level. We provide NMR and crystallographic evidence that the PucG protein from Bacillus subtilis catalyzes the transamination between an unstable intermediate ((S)-ureidoglycine) and the end product of purine catabolism (glyoxylate) to yield oxalurate and glycine. This activity enables soil and gut bacteria to use the animal purine waste as a source of carbon and nitrogen. The reaction catalyzed by (S)-ureidoglycine–glyoxylate aminotransferase (UGXT) illustrates a transamination sequence in which the same substrate provides both the amino group donor and, via its spontaneous decay, the amino group acceptor. Structural comparison and mutational analysis suggest a molecular rationale for the functional divergence between UGXT and peroxisomal alanine-glyoxylate aminotransferase, a fundamental enzyme for glyoxylate detoxification in humans.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Tipton, P.A. Urate to allantoin, specifically (S)-allantoin. Nat. Chem. Biol. 2, 124–125 (2006).
Vogels, G.D. & Van der Drift, C. Degradation of purines and pyrimidines by microorganisms. Bacteriol. Rev. 40, 403–468 (1976).
Todd, C.D. et al. Update on ureide degradation in legumes. J. Exp. Bot. 57, 5–12 (2006).
Werner, A.K., Romeis, T. & Witte, C.P. Ureide catabolism in Arabidopsis thaliana and Escherichia coli. Nat. Chem. Biol. 6, 19–21 (2010).
Serventi, F. et al. Chemical basis of nitrogen recovery through the ureide pathway: formation and hydrolysis of S-ureidoglycine in plants and bacteria. ACS Chem. Biol. 5, 203–214 (2010).
Gupta, S.C. & Dekker, E.E. Malyl-CoA formation in the NAD-, CoASH-, and alpha-ketoglutarate dehydrogenase-dependent oxidation of 2-keto-4-hydroxyglutarate. Possible coupled role of this reaction with 2-keto-4-hydroxyglutarate aldolase activity in a pyruvate-catalyzed cyclic oxidation of glyoxylate. J. Biol. Chem. 259, 10012–10019 (1984).
Zhang, X. et al. Crystal structure of alanine:glyoxylate aminotransferase and the relationship between genotype and enzymatic phenotype in primary hyperoxaluria type 1. J. Mol. Biol. 331, 643–652 (2003).
Schultz, A.C., Nygaard, P. & Saxild, H.H. Functional analysis of 14 genes that constitute the purine catabolic pathway in Bacillus subtilis and evidence for a novel regulon controlled by the PucR transcription activator. J. Bacteriol. 183, 3293–3302 (2001).
Salas, J.A., Johnstone, K. & Ellar, D.J. Role of uricase in the triggering of germination of Bacillus fastidiosus spores. Biochem. J. 229, 241–249 (1985).
Nakano, M.M. & Zuber, P. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52, 165–190 (1998).
Tam, N.K. et al. The intestinal life cycle of Bacillus subtilis and close relatives. J. Bacteriol. 188, 2692–2700 (2006).
Bowers, P.M., Cokus, S.J., Eisenberg, D. & Yeates, T.O. Use of logic relationships to decipher protein network organization. Science 306, 2246–2249 (2004).
Ramazzina, I. et al. Logical identification of an allantoinase analog (puuE) recruited from polysaccharide deacetylases. J. Biol. Chem. 283, 23295–23304 (2008).
Schneider, G., Kack, H. & Lindqvist, Y. The manifold of vitamin B6 dependent enzymes. Structure 8, R1–R6 (2000).
Han, Q. et al. Crystal structures of Aedes aegypti alanine glyoxylate aminotransferase. J. Biol. Chem. 281, 37175–37182 (2006).
Katsura, Y. et al. Crystal structure of a putative aspartate aminotransferase belonging to subgroup IV. Proteins 55, 487–492 (2004).
Rabinowitz, J.C. & Barker, H.A. Purine fermentation by Clostridium cylindrosporum. I. Tracer experiments on the fermentation of guanine. J. Biol. Chem. 218, 147–160 (1956).
Cellini, B., Bertoldi, M., Montioli, R., Paiardini, A. & Borri Voltattorni, C. Human wild-type alanine:glyoxylate aminotransferase and its naturally occurring G82E variant: functional properties and physiological implications. Biochem. J. 408, 39–50 (2007).
French, J.B. & Ealick, S.E. Biochemical and structural characterization of a ureidoglycine aminotransferase in the Klebsiella pneumoniae uric acid catabolic pathway. Biochemistry 49, 5975–5977 (2010).
Potrikus, C.J. & Breznak, J.A. Gut bacteria recycle uric acid nitrogen in termites: A strategy for nutrient conservation. Proc. Natl. Acad. Sci. USA 78, 4601–4605 (1981).
Singer, M.A. Do mammals, birds, reptiles and fish have similar nitrogen conserving systems? Comp. Biochem. Physiol. B 134, 543–558 (2003).
Hoa, N.T. et al. Characterization of Bacillus species used for oral bacteriotherapy and bacterioprophylaxis of gastrointestinal disorders. Appl. Environ. Microbiol. 66, 5241–5247 (2000).
Sorensen, L.B. & Levinson, D.J. Origin and extrarenal elimination of uric acid in man. Nephron 14, 7–20 (1975).
Kahn, K., Serfozo, P. & Tipton, P.A. Identification of the true product of the urate oxidase reaction. J. Am. Chem. Soc. 119, 5435–5442 (1997).
Gimisis, T. & Cismaş, C. Isolation, characterization, and independent synthesis of guanine oxidation products. Eur. J. Org. Chem. 2006, 1351–1378 (2006).
Percudani, R. & Peracchi, A. The B6 database: a tool for the description and classification of vitamin B6-dependent enzymatic activities and of the corresponding protein families. BMC Bioinformatics 10, 273 (2009).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Gouet, P., Courcelle, E., Stuart, D.I. & Metoz, F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 (1999).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Ye, Y. & Godzik, A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32, W582–W585 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Wilkins, M.R. et al. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 112, 531–552 (1999).
Kim, K., Park, J. & Rhee, S. Structural and functional basis for (S)-allantoin formation in the ureide pathway. J. Biol. Chem. 282, 23457–23464 (2007).
Schüttelkopf, A.W. & van Aalten, D.M. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 (2004).
Acknowledgements
We thank L. Borghi, A. Nouvenne and F. Albertini for discussions, and the staff of beamline XRD1 of Elettra, Trieste, for technical assistance during data collection.
Author information
Authors and Affiliations
Contributions
I.R. and R.C. performed experiments. L.C., R.B. and G.Z. performed the crystallographic studies. R.P. and A.P. designed experiments. R.P. conceived the study and wrote the paper with contributions from A.P. and G.Z. All authors analyzed data, discussed results and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods, Supplementary Tables 1–3 and Supplementary Figures 1–8 (PDF 1336 kb)
Rights and permissions
About this article
Cite this article
Ramazzina, I., Costa, R., Cendron, L. et al. An aminotransferase branch point connects purine catabolism to amino acid recycling. Nat Chem Biol 6, 801–806 (2010). https://doi.org/10.1038/nchembio.445
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.445
This article is cited by
-
E. coli allantoinase is activated by the downstream metabolic enzyme, glycerate kinase, and stabilizes the putative allantoin transporter by direct binding
Scientific Reports (2023)
-
In silico-guided metabolic engineering of Bacillus subtilis for efficient biosynthesis of purine nucleosides by blocking the key backflow nodes
Biotechnology for Biofuels and Bioproducts (2022)
-
Uric acid extrarenal excretion: the gut microbiome as an evident yet understated factor in gout development
Rheumatology International (2022)
-
Taxonomic variations in the gut microbiome of gout patients with and without tophi might have a functional impact on urate metabolism
Molecular Medicine (2021)
-
Identifying reaction modules in metabolic pathways: bioinformatic deduction and experimental validation of a new putative route in purine catabolism
BMC Systems Biology (2013)