Abstract
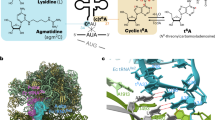
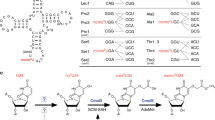
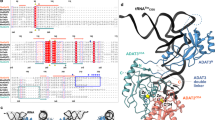
A modified base at the first (wobble) position of some tRNA anticodons is critical for deciphering the genetic code. In eukaryotes and eubacteria, AUA codons are decoded by tRNAsIle with modified bases pseudouridine (and/or inosine) and lysidine, respectively. The mechanism by which archaeal species translate AUA codons is unclear. We describe a polyamine-conjugated modified base, 2-agmatinylcytidine (agm2C or agmatidine), at the wobble position of archaeal tRNAIle that decodes AUA codons specifically. We demonstrate that archaeal cells use agmatine to synthesize agm2C of tRNAIle. We also identified a new enzyme, tRNAIle-agm2C synthetase (TiaS), that catalyzes agm2C formation in the presence of agmatine and ATP. Although agm2C is chemically similar to lysidine, TiaS constitutes a distinct class of enzyme from tRNAIle-lysidine synthetase (TilS), suggesting that the decoding systems evolved convergently across domains.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Yokoyama, S. & Nishimura, S. Modified nucleosides and codon recognition. in tRNA: Structure, Biosynthesis, and Function (eds. Söll, D. & RajBhandary, U.L.) 207–224 (American Society for Microbiology, Washington, DC, 1995).
Bjork, G. Biosynthesis and function of modified nucleosides. in tRNA: Structure, Biosynthesis, and Function (eds. Söll, D. & RajBhandary, U.L.) 165–205 (American Society for Microbiology, Washington, DC, 1995).
Suzuki, T. Biosynthesis and function of tRNA wobble modifications. in Topics in Current Genetics 24–69 (Springer-Verlag, New York, 2005).
Crick, F.H. Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555 (1966).
Senger, B., Auxilien, S., Englisch, U., Cramer, F. & Fasiolo, F. The modified wobble base inosine in yeast tRNAIle is a positive determinant for aminoacylation by isoleucyl-tRNA synthetase. Biochemistry 36, 8269–8275 (1997).
Muramatsu, T. et al. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J. Biol. Chem. 263, 9261–9267 (1988).
Muramatsu, T. et al. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336, 179–181 (1988).
Soma, A. et al. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell 12, 689–698 (2003).
Ikeuchi, Y. et al. molecular mechanism of lysidine synthesis that determines tRNA identity and codon recognition. Mol. Cell 19, 235–246 (2005).
Nakanishi, K. et al. Structural basis for translational fidelity ensured by transfer RNA lysidine synthetase. Nature 461, 1144–1148 (2009).
Suzuki, T. & Miyauchi, K. Discovery and characterization of tRNAIle lysidine synthetase (TilS). FEBS Lett. 584, 272–277 (2010).
Salowe, S.P., Wiltsie, J., Hawkins, J.C. & Sonatore, L.M. The catalytic flexibility of tRNAIle-lysidine synthetase can generate alternative tRNA substrates for isoleucyl-tRNA synthetase. J. Biol. Chem. 284, 9656–9662 (2009).
Edmonds, C.G. et al. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J. Bacteriol. 173, 3138–3148 (1991).
Gupta, R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 259, 9461–9471 (1984).
Kohrer, C. et al. Identification and characterization of a tRNA decoding the rare AUA codon in Haloarcula marismortui. RNA 14, 117–126 (2008).
Kanehisa, M. et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36, D480–D484 (2008).
Tatusov, R.L., Koonin, E.V. & Lipman, D.J. A genomic perspective on protein families. Science 278, 631–637 (1997).
Randau, L., Pearson, M. & Söll, D. The complete set of tRNA species in Nanoarchaeum equitans. FEBS Lett. 579, 2945–2947 (2005).
Elkins, J.G. et al. A korarchaeal genome reveals insights into the evolution of the Archaea. Proc. Natl. Acad. Sci. USA 105, 8102–8107 (2008).
Grosjean, H., Gaspin, C., Marck, C. & Decatur, W.A. & de Crecy-Lagard, V. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics 9, 470 (2008).
Groppa, M.D. & Benavides, M.P. Polyamines and abiotic stress: recent advances. Amino Acids 34, 35–45 (2008).
Terui, Y., Ohnuma, M., Hiraga, K., Kawashima, E. & Oshima, T. Stabilization of nucleic acids by unusual polyamines produced by an extreme thermophile, Thermus thermophilus. Biochem. J. 388, 427–433 (2005).
Tabor, C.W. & Tabor, H. Polyamines in microorganisms. Microbiol. Rev. 49, 81–99 (1985).
Fukuda, W., Morimoto, N., Imanaka, T. & Fujiwara, S. Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287, 113–120 (2008).
Ikeuchi, Y., Shigi, N., Kato, J., Nishimura, A. & Suzuki, T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 21, 97–108 (2006).
Noma, A., Kirino, Y., Ikeuchi, Y. & Suzuki, T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 25, 2142–2154 (2006).
Suzuki, T., Ikeuchi, Y., Noma, A., Suzuki, T. & Sakaguchi, Y. Mass spectrometric identification and characterization of RNA-modifying enzymes. Methods Enzymol. 425, 211–229 (2007).
Brock, T.D., Brock, K.M., Belly, R.T. & Weiss, R.L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch. Mikrobiol. 84, 54–68 (1972).
Mahapatra, A. et al. Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine. Mol. Microbiol. 59, 56–66 (2006).
Ogle, J.M., Murphy, F.V., Tarry, M.J. & Ramakrishnan, V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111, 721–732 (2002).
Acknowledgements
We are grateful to Y. Sakaguchi, T. Saigo, K. Nishikawa, S. Ohno and Y. Nomura for technical support and many fruitful discussions. Special thanks are due to Thermo Fischer Scientific for FT-MS analysis. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports, and Culture of Japan (to Tsutomu Suzuki, T.Y. and T.W.); by a Japan Society for the Promotion of Science Fellowship for Japanese Junior Scientists (to Y.I.); by a PRESTO program grant from Japan Science and Technology (to T.N.) and by a grant from the New Energy and Industrial Technology Development Organization (NEDO) (to Tsutomu Suzuki).
Author information
Authors and Affiliations
Contributions
Y.I. determined the chemical structure of agm2C. S.K. performed biochemical studies of agm2C and TiaS. Takeo Suzuki performed in vivo labeling of agm2C. T.N. performed expression and purification of TiaS. D.N. purified native tRNA under the supervision of T.Y. T.O. supported chemical synthesis of agm2C under the supervision of T.W. All authors discussed the results and commented on the manuscript. Tsutomu Suzuki designed and supervised all the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Methods and Supplementary Figures 1–9 (PDF 2310 kb)
Rights and permissions
About this article
Cite this article
Ikeuchi, Y., Kimura, S., Numata, T. et al. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat Chem Biol 6, 277–282 (2010). https://doi.org/10.1038/nchembio.323
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.323
This article is cited by
-
Genomic and transcriptomic dissection of Theionarchaea in marine ecosystem
Science China Life Sciences (2022)
-
The expanding world of tRNA modifications and their disease relevance
Nature Reviews Molecular Cell Biology (2021)
-
Probing conformational transitions towards mutagenic Watson–Crick-like G·T mismatches using off-resonance sugar carbon R1ρ relaxation dispersion
Journal of Biomolecular NMR (2020)
-
Identification of a radical SAM enzyme involved in the synthesis of archaeosine
Nature Chemical Biology (2019)
-
NAIL-MS reveals the repair of 2-methylthiocytidine by AlkB in E. coli
Nature Communications (2019)