Abstract
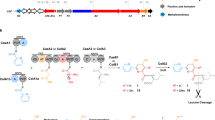
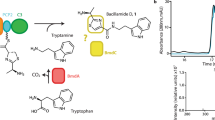
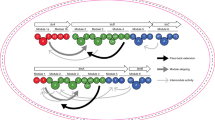
Despite containing an α-amino acid, the versatile cofactor S-adenosylmethionine (SAM) is not a known building block for nonribosomal peptide synthetase (NRPS) assembly lines. Here we report an unusual NRPS module from colibactin biosynthesis that uses SAM for amide bond formation and subsequent cyclopropanation. Our findings showcase a new use for SAM and reveal a novel biosynthetic route to a functional group that likely mediates colibactin's genotoxicity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Accession codes
References
Thibodeaux, C.J., Chang, W.C. & Liu, H.W. Chem. Rev. 112, 1681–1709 (2012).
Huang, S. Nat. Rev. Cancer 2, 469–476 (2002).
Broderick, J.B., Duffus, B.R., Duschene, K.S. & Shepard, E.M. Chem. Rev. 114, 4229–4317 (2014).
Walsh, C.T., O'Brien, R.V. & Khosla, C. Angew. Chem. Int. Ed. Engl. 52, 7098–7124 (2013).
Fischbach, M.A. & Walsh, C.T. Chem. Rev. 106, 3468–3496 (2006).
Nougayrède, J.P. et al. Science 313, 848–851 (2006).
Arthur, J.C. et al. Science 338, 120–123 (2012).
Vizcaino, M.I., Engel, P., Trautman, E. & Crawford, J.M. J. Am. Chem. Soc. 136, 9244–9247 (2014).
Brotherton, C.A., Wilson, M., Byrd, G. & Balskus, E.P. Org. Lett. 17, 1545–1548 (2015).
Bian, X.Y., Plaza, A., Zhang, Y.M. & Müller, R. Chem. Sci. 6, 3154–3160 (2015).
Vizcaino, M.I. & Crawford, J.M. Nat. Chem. 7, 411–417 (2015).
Li, Z.R. et al. ChemBioChem 16, 1715–1719 (2015).
Zha, L., Wilson, M.R., Brotherton, C.A. & Balskus, E.P. ACS Chem. Biol. 11, 1287–1295 (2016).
Li, Z.R. et al. Nat. Chem. Biol. 12, 773–775 (2016).
Ghosh, N., Sheldrake, H.M., Searcey, M. & Pors, K. Curr. Top. Med. Chem. 9, 1494–1524 (2009).
Tanasova, M. & Sturla, S.J. Chem. Rev. 112, 3578–3610 (2012).
Healy, A.R., Nikolayevskiy, H., Patel, J.R., Crawford, J.M. & Herzon, S.B. J. Am. Chem. Soc. 138, 15563–15570 (2016).
Brachmann, A.O. et al. Chem. Commun. (Camb.) 51, 13138–13141 (2015).
Yin, J., Lin, A.J., Golan, D.E. & Walsh, C.T. Nat. Protoc. 1, 280–285 (2006).
Dorrestein, P.C. et al. Biochemistry 45, 12756–12766 (2006).
Fontecave, M., Atta, M. & Mulliez, E. Trends Biochem. Sci. 29, 243–249 (2004).
Calderone, C.T., Kowtoniuk, W.E., Kelleher, N.L., Walsh, C.T. & Dorrestein, P.C. Proc. Natl. Acad. Sci. USA 103, 8977–8982 (2006).
Simunovic, V. & Müller, R. ChemBioChem 8, 1273–1280 (2007).
Stachelhaus, T., Mootz, H.D. & Marahiel, M.A. Chem. Biol. 6, 493–505 (1999).
Sundaram, S. & Hertweck, C. Curr. Opin. Chem. Biol. 31, 82–94 (2016).
Brotherton, C.A. & Balskus, E.P. J. Am. Chem. Soc. 135, 3359–3362 (2013).
Wenger, C.D., Phanstiel, D.H., Lee, M.V., Bailey, D.J. & Coon, J.J. Proteomics 11, 1064–1074 (2011).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Electrophoresis 20, 3551–3567 (1999).
Datsenko, K.A. & Wanner, B.L. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Bachmann, B.O. & Ravel, J. Methods Enzymol. 458, 181–217 (2009).
Sievers, F. et al. Mol. Syst. Biol. 7, 539 (2011).
Weber, T. et al. Nucleic Acids Res. 43, W237–W243 (2015).
Alva, V., Nam, S.Z., Soding, J. & Lupas, A.N. Nucleic Acids Res. 44, W410–W415 (2016).
Conti, E., Stachelhaus, T., Marahiel, M.A. & Brick, P. EMBO J. 16, 4174–4183 (1997).
Acknowledgements
We thank C. Brotherton and A. Sieg for help with cloning; J. May (Kahne lab, Harvard University, Cambridge, MA) for help with radiometric assays; M. McCallum for help with bioinformatic analyses; and P. Boudreau, C. Chittim, D. Kenny, H. Nakamura, and S. Peck for helpful discussions. L.Z., Y.J., M.R.W., and E.P.B. acknowledge financial support from National Cancer Institute (1R01CA208834-01), the Damon Runyon–Rachleff Innovation Award, and the Packard Fellowship for Science and Engineering. M.T.H. and N.L.K. were supported by Northwestern University and the US National Institutes of Health (GM 067725 and AT 009143). M.R.W. is supported by an American Cancer Society-New England Division Postdoctoral Fellowship (PF-16-122-01-CDD).
Author information
Authors and Affiliations
Contributions
L.Z. cloned, overexpressed, and purified colibactin biosynthetic enzymes and completed all enzymatic assays and bioinformatic analyses. Y.J. generated mutants used in the study and prepared culture extracts. M.T.H. analyzed enzymatic assays by protein mass spectrometry. M.R.W. prepared and characterized synthetic standards. J.X.W. helped to develop the analysis method for small-molecule LC–MS/MS. All coauthors collected and analyzed data. L.Z., M.T.H., N.L.K., and E.P.B. prepared and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–6 and Supplementary Figures 1–26 (PDF 7897 kb)
Supplementary Note
Preparation and characterization of synthetic standards 2 and 3 (PDF 1564 kb)
Rights and permissions
About this article
Cite this article
Zha, L., Jiang, Y., Henke, M. et al. Colibactin assembly line enzymes use S-adenosylmethionine to build a cyclopropane ring. Nat Chem Biol 13, 1063–1065 (2017). https://doi.org/10.1038/nchembio.2448
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2448
This article is cited by
-
Three-membered ring formation catalyzed by α-ketoglutarate-dependent nonheme iron enzymes
Journal of Natural Medicines (2024)
-
Pathogenic bacteria remodel central metabolic enzyme to build a cyclopropanol warhead
Nature Chemistry (2022)
-
Colibactin DNA-damage signature indicates mutational impact in colorectal cancer
Nature Medicine (2020)
-
Macrocyclic colibactin induces DNA double-strand breaks via copper-mediated oxidative cleavage
Nature Chemistry (2019)
-
Refining and expanding nonribosomal peptide synthetase function and mechanism
Journal of Industrial Microbiology and Biotechnology (2019)