Abstract
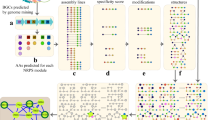
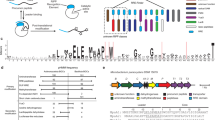
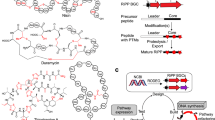
Ribosomally synthesized and post-translationally modified peptide (RiPP) natural products are attractive for genome-driven discovery and re-engineering, but limitations in bioinformatic methods and exponentially increasing genomic data make large-scale mining of RiPP data difficult. We report RODEO (Rapid ORF Description and Evaluation Online), which combines hidden-Markov-model-based analysis, heuristic scoring, and machine learning to identify biosynthetic gene clusters and predict RiPP precursor peptides. We initially focused on lasso peptides, which display intriguing physicochemical properties and bioactivities, but their hypervariability renders them challenging prospects for automated mining. Our approach yielded the most comprehensive mapping to date of lasso peptide space, revealing >1,300 compounds. We characterized the structures and bioactivities of six lasso peptides, prioritized based on predicted structural novelty, including one with an unprecedented handcuff-like topology and another with a citrulline modification exceptionally rare among bacteria. These combined insights significantly expand the knowledge of lasso peptides and, more broadly, provide a framework for future genome-mining efforts.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Primary accessions
GenBank/EMBL/DDBJ
Protein Data Bank
Referenced accessions
GenBank/EMBL/DDBJ
Protein Data Bank
References
Newman, D.J. & Cragg, G.M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016).
Winter, J.M., Behnken, S. & Hertweck, C. Genomics-inspired discovery of natural products. Curr. Opin. Chem. Biol. 15, 22–31 (2011).
Medema, M.H. et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 11, 625–631 (2015).
Weber, T. et al. antiSMASH 3.0—a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43, W237–W243 (2015).
Tietz, J.I. & Mitchell, D.A. Using genomics for natural product structure elucidation. Curr. Top. Med. Chem. 16, 1645–1694 (2016).
Stachelhaus, T., Mootz, H.D. & Marahiel, M.A. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6, 493–505 (1999).
Skinnider, M.A. et al. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 43, 9645–9662 (2015).
Cimermancic, P. et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158, 412–421 (2014).
Doroghazi, J.R. et al. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat. Chem. Biol. 10, 963–968 (2014).
Arnison, P.G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
Burkhart, B.J., Hudson, G.A., Dunbar, K.L. & Mitchell, D.A. A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 11, 564–570 (2015).
van Heel, A.J., de Jong, A., Montalbán-López, M., Kok, J. & Kuipers, O.P. BAGEL3: Automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res. 41, W448–W453 (2013).
Hegemann, J.D., Zimmermann, M., Xie, X. & Marahiel, M.A. Lasso peptides: an intriguing class of bacterial natural products. Acc. Chem. Res. 48, 1909–1919 (2015).
Al Toma, R.S. et al. Site-directed and global incorporation of orthogonal and isostructural noncanonical amino acids into the ribosomal lasso peptide capistruin. ChemBioChem 16, 503–509 (2015).
Pan, S.J., Rajniak, J., Maksimov, M.O. & Link, A.J. The role of a conserved threonine residue in the leader peptide of lasso peptide precursors. Chem. Commun. 48, 1880–1882 (2012).
Zong, C., Maksimov, M.O. & Link, A.J. Construction of lasso peptide fusion proteins. ACS Chem. Biol. 11, 61–68 (2016).
Kans, J. Entrez Direct: E-utilities on the UNIX Command Line (National Center for Biotechnology Information, 2010).
Finn, R.D. et al. HMMER web server: 2015 update. Nucleic Acids Res. 43 W1, W30–W38 (2015).
Finn, R.D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Maksimov, M.O., Pelczer, I. & Link, A.J. Precursor-centric genome-mining approach for lasso peptide discovery. Proc. Natl. Acad. Sci. USA 109, 15223–15228 (2012).
Libbrecht, M.W. & Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 16, 321–332 (2015).
Röttig, M. et al. NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 39, W362–W367 (2011).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Bailey, T.L., Johnson, J., Grant, C.E. & Noble, W.S. The MEME Suite. Nucleic Acids Res. 43, W39–W49 (2015).
Blin, K., Medema, M.H., Kottmann, R., Lee, S.Y. & Weber, T. The antiSMASH database, a comprehensive database of microbial secondary metabolite biosynthetic gene clusters. Nucleic Acids Res. 45, D555–D559 (2017).
Skinnider, M.A. et al. Genomic charting of ribosomally synthesized natural product chemical space facilitates targeted mining. Proc. Natl. Acad. Sci. USA 113, E6343–E6351 (2016).
Gerlt, J.A. et al. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): a web tool for generating protein sequence similarity networks. Biochim. Biophys. Acta 1854, 1019–1037 (2015).
Maksimov, M.O., Pan, S.J. & James Link, A. Lasso peptides: structure, function, biosynthesis, and engineering. Nat. Prod. Rep. 29, 996–1006 (2012).
Zimmermann, M., Hegemann, J.D., Xie, X.L. & Marahiel, M.A. Characterization of caulonodin lasso peptides revealed unprecedented N-terminal residues and a precursor motif essential for peptide maturation. Chem. Sci. 5, 4032–4043 (2014).
Metelev, M. et al. Structure, bioactivity, and resistance mechanism of streptomonomicin, an unusual lasso peptide from an understudied halophilic actinomycete. Chem. Biol. 22, 241–250 (2015).
Hegemann, J.D. et al. The ring residue proline 8 is crucial for the thermal stability of the lasso peptide caulosegnin II. Mol. Biosyst. 12, 1106–1109 (2016).
UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Hopf, T.A. et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. eLife 3, e03430 (2014).
Ovchinnikov, S., Kamisetty, H. & Baker, D. Robust and accurate prediction of residue-residue interactions across protein interfaces using evolutionary information. eLife 3, e02030 (2014).
Balakrishnan, S., Kamisetty, H., Carbonell, J.G., Lee, S.I. & Langmead, C.J. Learning generative models for protein fold families. Proteins 79, 1061–1078 (2011).
Koehnke, J. et al. Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat. Chem. Biol. 11, 558–563 (2015).
Maksimov, M.O. & Link, A.J. Discovery and characterization of an isopeptidase that linearizes lasso peptides. J. Am. Chem. Soc. 135, 12038–12047 (2013).
Weber, W., Fischli, W., Hochuli, E., Kupfer, E. & Weibel, E.K. Anantin--a peptide antagonist of the atrial natriuretic factor (ANF). I. Producing organism, fermentation, isolation and biological activity. J. Antibiot. 44, 164–171 (1991).
Xie, X. & Marahiel, M.A. NMR as an effective tool for the structure determination of lasso peptides. ChemBioChem 13, 621–625 (2012).
Ogawa, T. et al. RES-701-2, -3 and -4, novel and selective endothelin type B receptor antagonists produced by Streptomyces sp. I. Taxonomy of producing strains, fermentation, isolation, and biochemical properties. J. Antibiot. 48, 1213–1220 (1995).
Gavrish, E. et al. Lassomycin, a ribosomally synthesized cyclic peptide, kills mycobacterium tuberculosis by targeting the ATP-dependent protease ClpC1P1P2. Chem. Biol. 21, 509–518 (2014).
Goulas, T. et al. Structure and mechanism of a bacterial host-protein citrullinating virulence factor, Porphyromonas gingivalis peptidylarginine deiminase. Sci. Rep. 5, 11969 (2015).
Markowitz, V.M. et al. IMG 4 version of the integrated microbial genomes comparative analysis system. Nucleic Acids Res. 42, D560–D567 (2014).
Fong, C., Rohmer, L., Radey, M., Wasnick, M. & Brittnacher, M.J. PSAT: a web tool to compare genomic neighborhoods of multiple prokaryotic genomes. BMC Bioinformatics 9, 170 (2008).
Molloy, E.M., Tietz, J.I., Blair, P.M. & Mitchell, D.A. Biological characterization of the hygrobafilomycin antibiotic JBIR-100 and bioinformatic insights into the hygrolide family of natural products. Bioorg. Med. Chem. 24, 6276–6290 (2016).
Li, Y. et al. Characterization of sviceucin from streptomyces provides insight into enzyme exchangeability and disulfide bond formation in lasso peptides. ACS Chem. Biol. 10, 2641–2649 (2015).
McGraw, W.T., Potempa, J., Farley, D. & Travis, J. Purification, characterization, and sequence analysis of a potential virulence factor from Porphyromonas gingivalis, peptidylarginine deiminase. Infect. Immun. 67, 3248–3256 (1999).
Gabarrini, G. et al. The peptidylarginine deiminase gene is a conserved feature of Porphyromonas gingivalis. Sci. Rep. 5, 13936 (2015).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Hildebrand, A., Remmert, M., Biegert, A. & Söding, J. Fast and accurate automatic structure prediction with HHpred. Proteins 77 (Suppl. 9), 128–132 (2009).
Letunic, I. & Bork, P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 39, W475–W478 (2011).
Su, G., Morris, J.H., Demchak, B. & Bader, G.D. Biological network exploration with Cytoscape 3. Curr. Protoc. Bioinformatics 47, 8.13.1–8.1324 (2014).
Kohl, M., Wiese, S. & Warscheid, B. Cytoscape: software for visualization and analysis of biological networks. Methods Mol. Biol. 696, 291–303 (2011).
Krzywinski, M. et al. Circos: an information aesthetic for comparative genomics. Genome Res. 19, 1639–1645 (2009).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Eliot, A.C. et al. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem. Biol. 15, 765–770 (2008).
Schwieters, C.D., Kuszewski, J.J. & Clore, G.M. Using Xplor-NIH for NMR molecular structure determination. Prog. Nucl. Magn. Reson. Spectrosc. 48, 47–62 (2006).
Acknowledgements
We thank L. Zhu (University of Illinois Urbana–Champaign) and B. Ramirez (University of Illinois Chicago) for NMR assistance. We acknowledge C. Cox, K. Choe (University of Illinois Urbana–Champaign), and N. Tietz for valuable computational and programming input. This work was supported in part by a NIH Director's New Innovator Award Program (DP2 OD008463 to D.A.M.), the David and Lucile Packard Fellowship for Science and Engineering (to D.A.M.), the Robert C. and Carolyn J. Springborn Endowment (to J.I.T.), and ACS Division of Medicinal Chemistry Predoctoral Fellowships (to P.M.B. and J.I.T.). C.J.S. is a member of the NIH Chemistry–Biology Interface Training Program (Grant NRSA 1-T32-GM070421). The Bruker UltrafleXtreme MALDI TOF/TOF mass spectrometer was purchased in part with a grant from the National Center for Research Resources, National Institutes of Health (S10 RR027109 A). The 900 MHz NMR spectrometer was purchased with funds provided by GM068944.
Author information
Authors and Affiliations
Contributions
J.I.T. and C.J.S. contributed equally to this work. Experiments were designed by J.I.T., C.J.S. and D.A.M. and were performed by J.I.T., C.J.S., T.M., P.M.B., H.-C.T. and U.I.Z. J.I.T., P.S.P. and C.J.S. wrote the code. The manuscript was written by J.I.T., C.J.S., and D.A.M.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–11, Supplementary Figures 1–15 and Supplementary Notes 1–3 (PDF 9591 kb)
Supplementary Dataset 1
Spreadsheet containing all RODEO-derived information on lasso peptide precursors, BGCs, co-occurrence. (XLSX 14569 kb)
Supplementary Dataset 2
Cytoscape-readable file containing individual sequence similarity networks for the lasso precursors, lasso cyclase, leader peptidase, and RRE (PqqD) proteins. (ZIP 20183 kb)
Supplementary Dataset 3
High-resolution PDF showing a Circos diagram of all RODEO-identified lasso peptide BGCs with annotations indicating co-occurring Pfam information. (PDF 28576 kb)
Rights and permissions
About this article
Cite this article
Tietz, J., Schwalen, C., Patel, P. et al. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat Chem Biol 13, 470–478 (2017). https://doi.org/10.1038/nchembio.2319
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2319
This article is cited by
-
The Complete Genomic Sequence of Microbial Transglutaminase Producer, Streptomyces mobaraensis DSM40587
Biochemical Genetics (2024)
-
Systematic mining of the human microbiome identifies antimicrobial peptides with diverse activity spectra
Nature Microbiology (2023)
-
Structure of lasso peptide epimerase MslH reveals metal-dependent acid/base catalytic mechanism
Nature Communications (2023)
-
Genome mining unveils a class of ribosomal peptides with two amino termini
Nature Communications (2023)
-
Construction of a prognostic prediction model for renal clear cell carcinoma combining clinical traits
Scientific Reports (2023)