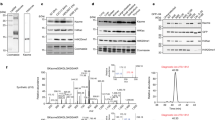
Abstract
ADP-ribosylation (ADPr) is a biologically and clinically important post-translational modification, but little is known about the amino acids it targets on cellular proteins. Here we present a proteomic approach for direct in vivo identification and quantification of ADPr sites on histones. We have identified 12 unique ADPr sites in human osteosarcoma cells and report serine ADPr as a new type of histone mark that responds to DNA damage.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Feng, F.Y., de Bono, J.S., Rubin, M.A. & Knudsen, K.E. Mol. Cell 58, 925–934 (2015).
Daniels, C.M., Ong, S.E. & Leung, A.K. Mol. Cell 58, 911–924 (2015).
Hottiger, M.O. Annu. Rev. Biochem. 84, 227–263 (2015).
Huang, H., Lin, S., Garcia, B.A. & Zhao, Y. Chem. Rev. 115, 2376–2418 (2015).
Tan, M. et al. Cell 146, 1016–1028 (2011).
Wiśniewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Nat. Methods 6, 359–362 (2009).
Rodriguez-Collazo, P., Leuba, S.H. & Zlatanova, J. Nucleic Acids Res. 37, e81 (2009).
Jungmichel, S. et al. Mol. Cell 52, 272–285 (2013).
Daniels, C.M., Ong, S.E. & Leung, A.K. J. Proteome Res. 13, 3510–3522 (2014).
Palazzo, L. et al. Biochem. J. 468, 293–301 (2015).
Daniels, C.M., Thirawatananond, P., Ong, S.E., Gabelli, S.B. & Leung, A.K. Sci. Rep. 5, 18271 (2015).
Rosenthal, F., Nanni, P., Barkow-Oesterreicher, S. & Hottiger, M.O. J. Proteome Res. 14, 4072–4079 (2015).
Steentoft, C. et al. Nat. Methods 8, 977–982 (2011).
Cox, J. & Mann, M. Nat. Biotechnol. 26, 1367–1372 (2008).
Messner, S. et al. Nucleic Acids Res. 38, 6350–6362 (2010).
Cervantes-Laurean, D., Loflin, P.T., Minter, D.E., Jacobson, E.L. & Jacobson, M.K. J. Biol. Chem. 270, 7929–7936 (1995).
Messner, S. & Hottiger, M.O. Trends Cell Biol. 21, 534–542 (2011).
Ong, S.E. et al. Mol. Cell. Proteomics 1, 376–386 (2002).
D'Amours, D., Desnoyers, S., D'Silva, I. & Poirier, G.G. Biochem. J. 342, 249–268 (1999).
Moremen, K.W., Tiemeyer, M. & Nairn, A.V. Nat. Rev. Mol. Cell Biol. 13, 448–462 (2012).
Wang, F. & Higgins, J.M. Trends Cell Biol. 23, 175–184 (2013).
Affar, E.B. et al. Anal. Biochem. 259, 280–283 (1998).
Slade, D. et al. Nature 477, 616–620 (2011).
Kleine, H. et al. Mol. Cell 32, 57–69 (2008).
Rappsilber, J., Ishihama, Y. & Mann, M. Anal. Chem. 75, 663–670 (2003).
Kelstrup, C.D., Young, C., Lavallee, R., Nielsen, M.L. & Olsen, J.V. J. Proteome Res. 11, 3487–3497 (2012).
Cox, J. et al. J. Proteome Res. 10, 1794–1805 (2011).
Hengel, S.M. & Goodlett, D.R. Int. J. Mass Spectrom. 312, 114–121 (2012).
Acknowledgements
Many thanks to A. Zhiteneva, W. Earnshaw (University of Edinburgh) and M. Tatham (University of Dundee) for comments on the manuscript. Chicken bursal lymphoma DT40 cells were a gift from W. Earnshaw (Wellcome Trust Centre for Cell Biology, University of Edinburgh, Edinburgh, UK). pGEX-4T1 GST-PARP10cd plasmid was a gift from B. Lüscher (Department of Biochemistry and Molecular Biology, RWTH Aachen University, Aachen, Germany). This work was funded by Deutsche Forschungsgemeinschaft (Cellular Stress Responses in Aging-Associated Diseases) (grant EXC 229 to I.M.) and the European Union's Horizon 2020 research and innovation program (Marie Sklodowska-Curie grant agreement no. 657501 to J.J.B. and I.M.). The work in the Ahel laboratory is funded by the Wellcome Trust (grant 101794) and the European Research Council (grant 281739).
Author information
Authors and Affiliations
Contributions
I.M., O.L. and J.J.B. designed research. O.L. and J.J.B. performed experiments and acquired and analyzed MS data. T.C. optimized MS methods and contributed to the acquisition and analysis of MS data. Q.Z. performed histone purification and digestion experiments. I. Atanassov performed bioinformatics analyses. A.S. performed immuno-slot blot experiments. L.P. and R.Z. performed protein purification and in vitro assays. I. Ahel and R.Z. contributed to the histone purification experiments and supporting studies. I.M. analyzed data and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–11, Supplementary Tables 1–2 and Supplementary Note. (PDF 16867 kb)
Supplementary Dataset 1
SILAC-based quantification of histone marks: DNA damage and olaparib experiments. (XLSX 69 kb)
Rights and permissions
About this article
Cite this article
Leidecker, O., Bonfiglio, J., Colby, T. et al. Serine is a new target residue for endogenous ADP-ribosylation on histones. Nat Chem Biol 12, 998–1000 (2016). https://doi.org/10.1038/nchembio.2180
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2180
This article is cited by
-
PARP2 promotes Break Induced Replication-mediated telomere fragility in response to replication stress
Nature Communications (2024)
-
High-throughput screening assay for PARP-HPF1 interaction inhibitors to affect DNA damage repair
Scientific Reports (2024)
-
Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling
Nature Communications (2023)
-
HPF1-dependent histone ADP-ribosylation triggers chromatin relaxation to promote the recruitment of repair factors at sites of DNA damage
Nature Structural & Molecular Biology (2023)
-
Regulation of Rad52-dependent replication fork recovery through serine ADP-ribosylation of PolD3
Nature Communications (2023)