Abstract
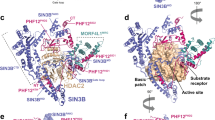
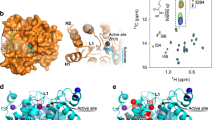
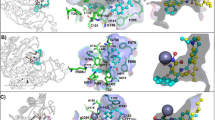
Histone deacetylase 6 (HDAC6) is a critical target for drug design because of its role in oncogenic transformation and cancer metastasis, and is unique among all histone deacetylases in that it contains tandem catalytic domains designated CD1 and CD2. We now report the crystal structures of CD2 from Homo sapiens HDAC6 and of CD1 and CD2 from Danio rerio HDAC6. We correlated these structures with activity measurements using 13 different substrates. The catalytic activity of CD2 from both species exhibited broad substrate specificity, whereas that of CD1 was highly specific for substrates bearing C-terminal acetyllysine residues. Crystal structures of substrate complexes yielded unprecedented snapshots of the catalytic mechanism. Additionally, crystal structures of complexes with eight different inhibitors, including belinostat and panobinostat (currently used in cancer chemotherapy), the macrocyclic tetrapeptide HC toxin, and the HDAC6-specific inhibitor N-hydroxy-4-(2-((2-hydroxyethyl)(phenyl)amino)-2-oxoethyl)benzamide, revealed surprising new insight regarding changes in Zn2+ coordination and isozyme-specific inhibition.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kouzarides, T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19, 1176–1179 (2000).
Norvell, A. & McMahon, S.B. Rise of the rival. Science 327, 964–965 (2010).
Choudhary, C. et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 (2009).
Berger, S.L., Kouzarides, T., Shiekhattar, R. & Shilatifard, A. An operational definition of epigenetics. Genes Dev. 23, 781–783 (2009).
Delcuve, G.P., Khan, D.H. & Davie, J.R. Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors. Clin. Epigenetics 4, 5 (2012).
Choudhary, C., Weinert, B.T., Nishida, Y., Verdin, E. & Mann, M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat. Rev. Mol. Cell Biol. 15, 536–550 (2014).
Zhao, S. et al. Regulation of cellular metabolism by protein lysine acetylation. Science 327, 1000–1004 (2010).
Wang, Q. et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 327, 1004–1007 (2010).
Verdin, E. & Ott, M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015).
Marmorstein, R. & Zhou, M.-M. Writers and readers of histone acetylation: structure, mechanism, and inhibition. Cold Spring Harb. Perspect. Biol. 6, a018762 (2014).
West, A.C. & Johnstone, R.W. New and emerging HDAC inhibitors for cancer treatment. J. Clin. Invest. 124, 30–39 (2014).
Ma, N. et al. Selective histone deacetylase inhibitors with anticancer activity. Curr. Top. Med. Chem. 16, 415–426 (2016).
Dokmanovic, M., Clarke, C. & Marks, P.A. Histone deacetylase inhibitors: overview and perspectives. Mol. Cancer Res. 5, 981–989 (2007).
Arrowsmith, C.H., Bountra, C., Fish, P.V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012).
Falkenberg, K.J. & Johnstone, R.W. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat. Rev. Drug Discov. 13, 673–691 (2014).
Gregoretti, I.V., Lee, Y.M. & Goodson, H.V. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 338, 17–31 (2004).
Lombardi, P.M., Cole, K.E., Dowling, D.P. & Christianson, D.W. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 21, 735–743 (2011).
Yuan, H. & Marmorstein, R. Structural basis for sirtuin activity and inhibition. J. Biol. Chem. 287, 42428–42435 (2012).
Grozinger, C.M., Hassig, C.A. & Schreiber, S.L. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. USA 96, 4868–4873 (1999).
Ouyang, H. et al. Protein aggregates are recruited to aggresome by histone deacetylase 6 via unanchored ubiquitin C termini. J. Biol. Chem. 287, 2317–2327 (2012).
Kawaguchi, Y. et al. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 (2003).
Zhang, M. et al. HDAC6 deacetylates and ubiquitinates MSH2 to maintain proper levels of MutSα. Mol. Cell 55, 31–46 (2014).
Bertos, N.R. et al. Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. J. Biol. Chem. 279, 48246–48254 (2004).
Hubbert, C. et al. HDAC6 is a microtubule-associated deacetylase. Nature 417, 455–458 (2002).
Szyk, A. et al. Molecular basis for age-dependent microtubule acetylation by tubulin acetyltransferase. Cell 157, 1405–1415 (2014).
Kovacs, J.J. et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell 18, 601–607 (2005).
Cohen, T.J. et al. The acetylation of tau inhibits its function and promotes pathological tau aggregation. Nat. Commun. 2, 252 (2011).
Min, S.W. et al. Acetylation of tau inhibits its degradation and contributes to tauopathy. Neuron 67, 953–966 (2010).
Haggarty, S.J., Koeller, K.M., Wong, J.C., Grozinger, C.M. & Schreiber, S.L. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proc. Natl. Acad. Sci. USA 100, 4389–4394 (2003).
Zou, H., Wu, Y., Navre, M. & Sang, B.-C. Characterization of the two catalytic domains in histone deacetylase 6. Biochem. Biophys. Res. Commun. 341, 45–50 (2006).
Zhang, Y., Gilquin, B., Khochbin, S. & Matthias, P. Two catalytic domains are required for protein deacetylation. J. Biol. Chem. 281, 2401–2404 (2006).
Schreiber, S.L. & Bernstein, B.E. Signaling network model of chromatin. Cell 111, 771–778 (2002).
Gantt, S.M. et al. General base-general acid catalysis in human histone deacetylase 8. Biochemistry 55, 820–832 (2016).
Pauling, L. Molecular architecture and biological reactions. Chem. Eng. News 24, 1375–1377 (1946).
Decroos, C., Bowman, C.M. & Christianson, D.W. Synthesis and evaluation of N8-acetylspermidine analogues as inhibitors of bacterial acetylpolyamine amidohydrolase. Bioorg. Med. Chem. 21, 4530–4540 (2013).
Liang, T.-C. & Abeles, R.H. Complex of α-chymotrypsin and N-acetyl-L-leucyl-L-phenylalanyl trifluoromethyl ketone: structural studies with NMR spectroscopy. Biochemistry 26, 7603–7608 (1987).
Vannini, A. et al. Substrate binding to histone deacetylases as shown by the crystal structure of the HDAC8-substrate complex. EMBO Rep. 8, 879–884 (2007).
Dowling, D.P., Gantt, S.L., Gattis, S.G., Fierke, C.A. & Christianson, D.W. Structural studies of human histone deacetylase 8 and its site-specific variants complexed with substrate and inhibitors. Biochemistry 47, 13554–13563 (2008).
Brosch, G., Ransom, R., Lechner, T., Walton, J.D. & Loidl, P. Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7, 1941–1950 (1995).
Furumai, R. et al. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc. Natl. Acad. Sci. USA 98, 87–92 (2001).
Christianson, D.W. & Lipscomb, W.N. Binding of a possible transition state analogue to the active site of carboxypeptidase A. Proc. Natl. Acad. Sci. USA 82, 6840–6844 (1985).
Christianson, D.W., David, P.R. & Lipscomb, W.N. Mechanism of carboxypeptidase A: hydration of a ketonic substrate analogue. Proc. Natl. Acad. Sci. USA 84, 1512–1515 (1987).
Lee, J.H. et al. Development of a histone deacetylase 6 inhibitor and its biological effects. Proc. Natl. Acad. Sci. USA 110, 15704–15709 (2013).
Matthews, B.W. Structural basis of the action of thermolysin and related zinc peptidases. Acc. Chem. Res. 21, 333–340 (1988).
Christianson, D.W. & Lipscomb, W.N. Carboxypeptidase A. Acc. Chem. Res. 22, 62–69 (1989).
Fischle, W. et al. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem. 274, 11713–11720 (1999).
Center, R.J. et al. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 7, 1612–1619 (1998).
Smyth, D.R., Mrozkiewicz, M.K., McGrath, W.J., Listwan, P. & Kobe, B. Crystal structures of fusion proteins with large-affinity tags. Protein Sci. 12, 1313–1322 (2003).
Decroos, C. et al. Compromised structure and function of HDAC8 mutants identified in Cornelia de Lange Syndrome spectrum disorders. ACS Chem. Biol. 9, 2157–2164 (2014).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
McCoy, A.J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Bürli, R.W. et al. Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease. J. Med. Chem. 56, 9934–9954 (2013).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Chen, V.B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Cheng, Y. & Prusoff, W.H. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol. 22, 3099–3108 (1973).
Hildmann, C. et al. A new amidohydrolase from Bordetella or Alcaligenes strain FB188 with similarities to histone deacetylases. J. Bacteriol. 186, 2328–2339 (2004).
Kim, B., Pithadia, A.S. & Fierke, C.A. Kinetics and thermodynamics of metal-binding to histone deacetylase 8. Protein Sci. 24, 354–365 (2015).
Acknowledgements
We thank R. Marmorstein, B.S. Cooperman, X. Li, and M. Chen for helpful discussions, the National Institutes of Health (NIH) for grant GM49758 to D.W.C. in support of this research, and V. Stojanoff, D. Neau, and J. Nix for assistance with synchrotron data collection. For synchrotron access, we are grateful to the Northeastern Collaborative Access Team beamline 24-ID-E at the Advanced Photon Source, Argonne National Laboratory; the Structural Molecular Biology Program beamline 14-1 at the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory; and the Molecular Biology Consortium beamline 4.2.2 at the Advanced Light Source, Lawrence Berkeley National Laboratory. Finally, D.W.C. thanks the Radcliffe Institute for Advanced Study at Harvard University for the Elizabeth S. and Richard M. Cashin Fellowship.
Author information
Authors and Affiliations
Contributions
Y.H. and D.W.C. designed the project. Y.H. performed experiments. Y.H. and D.W.C. interpreted experimental results and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–5 and Supplementary Figures 1–13. (PDF 15712 kb)
Rights and permissions
About this article
Cite this article
Hai, Y., Christianson, D. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat Chem Biol 12, 741–747 (2016). https://doi.org/10.1038/nchembio.2134
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2134
This article is cited by
-
Pharmacophore-based virtual screening of ZINC database, molecular modeling and designing new derivatives as potential HDAC6 inhibitors
Molecular Diversity (2023)
-
Phenolic compounds as histone deacetylase inhibitors: binding propensity and interaction insights from molecular docking and dynamics simulations
Amino Acids (2023)
-
Exploring new histone deacetylase 6 inhibitors and their effects on reversing the α-tubulin deacetylation and cell morphology changes caused by methamphetamine
Archives of Pharmacal Research (2023)
-
Three-dimensional quantitative structural-activity relationship and molecular dynamics study of multivariate substituted 4-oxyquinazoline HDAC6 inhibitors
Molecular Diversity (2023)
-
HDAC6 score: to treat or not to treat?
Nature Cancer (2022)