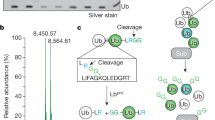
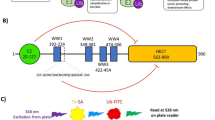
Abstract
E3 ligases represent an important class of enzymes, yet there are currently no chemical probes for profiling their activity. We develop a new class of activity-based probe by re-engineering a ubiquitin-charged E2 conjugating enzyme and demonstrate the utility of these probes by profiling the transthiolation activity of the RING-in-between-RING (RBR) E3 ligase parkin in vitro and in cellular extracts. Our study provides valuable insight into the roles, and cellular hierarchy, of distinct phosphorylation events in parkin activation. We also profile parkin mutations associated with patients with Parkinson's disease and demonstrate that they mediate their effect largely by altering transthiolation activity. Furthermore, our probes enable direct and quantitative measurement of endogenous parkin activity, revealing that endogenous parkin is activated in neuronal cell lines (≥75%) in response to mitochondrial depolarization. This new technology also holds promise as a novel biomarker of PINK1-parkin signaling, as demonstrated by its compatibility with samples derived from individuals with Parkinson's disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 (2014).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Berndsen, C.E. & Wolberger, C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 21, 301–307 (2014).
Kitada, T. et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 392, 605–608 (1998).
Wenzel, D.M., Lissounov, A., Brzovic, P.S. & Klevit, R.E. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011).
Pickrell, A.M. & Youle, R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron 85, 257–273 (2015).
Chaugule, V.K. et al. Autoregulation of parkin activity through its ubiquitin-like domain. EMBO J. 30, 2853–2867 (2011).
Trempe, J.F. et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science 340, 1451–1455 (2013).
Riley, B.E. et al. Structure and function of parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat. Commun. 4, 1982 (2013).
Wauer, T. & Komander, D. Structure of the human parkin ligase domain in an autoinhibited state. EMBO J. 32, 2099–2112 (2013).
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014).
Kazlauskaite, A. et al. Phosphorylation of parkin at Serine65 is essential for activation: elaboration of a Miro1 substrate-based assay of parkin E3 ligase activity. Open Biol. 4, 130213 (2014).
Kane, L.A. et al. PINK1 phosphorylates ubiquitin to activate parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014).
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014).
Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2, 120080 (2012).
Shiba-Fukushima, K. et al. PINK1-mediated phosphorylation of the parkin ubiquitin-like domain primes mitochondrial translocation of parkin and regulates mitophagy. Sci. Rep. 2, 1002 (2012).
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014).
Ordureau, A. et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl. Acad. Sci. USA 112, 6637–6642 (2015).
Okatsu, K. et al. Phosphorylated ubiquitin chain is the genuine parkin receptor. J. Cell Biol. 209, 111–128 (2015).
Kazlauskaite, A. et al. Binding to serine 65-phosphorylated ubiquitin primes parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 16, 939–954 (2015).
Wauer, T., Simicek, M., Schubert, A. & Komander, D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature 524, 370–374 (2015).
Kazlauskaite, A. & Muqit, M.M. PINK1 and parkin – mitochondrial interplay between phosphorylation and ubiquitylation in parkinson's disease. FEBS J. 282, 215–223 (2015).
Lazarou, M. et al. PINK1 drives parkin self-association and HECT-like E3 activity upstream of mitochondrial binding. J. Cell Biol. 200, 163–172 (2013).
Matsuda, N. et al. PINK1 stabilized by mitochondrial depolarization recruits parkin to damaged mitochondria and activates latent parkin for mitophagy. J. Cell Biol. 189, 211–221 (2010).
Cravatt, B.F., Wright, A.T. & Kozarich, J.W. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 77, 383–414 (2008).
Stanley, M. et al. Orthogonal thiol functionalization at a single atomic center for profiling transthiolation activity of E1 activating enzymes. ACS Chem. Biol. 10, 1542–1554 (2015).
Borodovsky, A. et al. A novel active site-directed probe specific for deubiquitylating enzymes reveals proteasome association of USP14. EMBO J. 20, 5187–5196 (2001).
Love, K.R., Pandya, R.K., Spooner, E. & Ploegh, H.L. Ubiquitin C-terminal electrophiles are activity-based probes for identification and mechanistic study of ubiquitin conjugating machinery. ACS Chem. Biol. 4, 275–287 (2009).
Smit, J.J. & Sixma, T.K. RBR E3-ligases at work. EMBO Rep. 15, 142–154 (2014).
Komander, D., Clague, M.J. & Urbé, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 (2009).
Olsen, S.K., Capili, A.D., Lu, X., Tan, D.S. & Lima, C.D. Active site remodelling accompanies thioester bond formation in the SUMO E1. Nature 463, 906–912 (2010).
McGouran, J.F., Gaertner, S.R., Altun, M., Kramer, H.B. & Kessler, B.M. Deubiquitinating enzyme specificity for ubiquitin chain topology profiled by di-ubiquitin activity probes. Chem. Biol. 20, 1447–1455 (2013).
Mulder, M.P., El Oualid, F., ter Beek, J. & Ovaa, H. A native chemical ligation handle that enables the synthesis of advanced activity-based probes: diubiquitin as a case study. ChemBioChem 15, 946–949 (2014).
Haj-Yahya, N. et al. Dehydroalanine-based diubiquitin activity probes. Org. Lett. 16, 540–543 (2014).
Li, G., Liang, Q., Gong, P., Tencer, A.H. & Zhuang, Z. Activity-based diubiquitin probes for elucidating the linkage specificity of deubiquitinating enzymes. Chem. Commun. (Camb.) 50, 216–218 (2014).
Borodovsky, A. et al. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9, 1149–1159 (2002).
Niphakis, M.J. & Cravatt, B.F. Enzyme inhibitor discovery by activity-based protein profiling. Annu. Rev. Biochem. 83, 341–377 (2014).
Nguyen, D.P. et al. Genetic encoding and labeling of aliphatic azides and alkynes in recombinant proteins via a pyrrolysyl-tRNA Synthetase/tRNA(CUA) pair and click chemistry. J. Am. Chem. Soc. 131, 8720–8721 (2009).
Weissman, A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2, 169–178 (2001).
Lechtenberg, B.C. et al. Structure of a HOIP/E2∼ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature 529, 546–550 (2016).
Sarraf, S.A. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 (2013).
Kamadurai, H.B. et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B∼ubiquitin-HECT(NEDD4L) complex. Mol. Cell 36, 1095–1102 (2009).
Stieglitz, B., Morris-Davies, A.C., Koliopoulos, M.G., Christodoulou, E. & Rittinger, K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 13, 840–846 (2012).
Lin, D.Y., Diao, J., Zhou, D. & Chen, J. Biochemical and structural studies of a HECT-like ubiquitin ligase from Escherichia coli O157:H7. J. Biol. Chem. 286, 441–449 (2011).
Ekkebus, R. et al. On terminal alkynes that can react with active-site cysteine nucleophiles in proteases. J. Am. Chem. Soc. 135, 2867–2870 (2013).
Lazarou, M. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 (2015).
Kumar, A. et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 34, 2506–2521 (2015).
Sauvé, V. et al. A Ubl/ubiquitin switch in the activation of parkin. EMBO J. 34, 2492–2505 (2015).
Okatsu, K. et al. A dimeric PINK1-containing complex on depolarized mitochondria stimulates parkin recruitment. J. Biol. Chem. 288, 36372–36384 (2013).
Lai, Y.C. et al. Phosphoproteomic screening identifies Rab GTPases as novel downstream targets of PINK1. EMBO J. 34, 2840–2861 (2015).
Katrun, P. & Chiampanichayakul, S.PhI. (OAc)2/KI-mediated reaction of aryl sulfinates with alkenes, alkynes, and α,β-unsaturated carbonyl compounds: synthesis of vinyl sulfones and β-iodovinyl sulfones. Eur. J. Org. Chem. 29, 5633–5641 (2010).
Dadová, J. et al. Vinylsulfonamide and acrylamide modification of DNA for cross-linking with proteins. Angew. Chem. Int. Edn. Engl. 52, 10515–10518 (2013).
Virdee, S., Ye, Y., Nguyen, D.P., Komander, D. & Chin, J.W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 6, 750–757 (2010).
Cui, C., Zhao, W., Chen, J., Wang, J. & Li, Q. Elimination of in vivo cleavage between target protein and intein in the intein-mediated protein purification systems. Protein Expr. Purif. 50, 74–81 (2006).
El Oualid, F. et al. Chemical synthesis of ubiquitin, ubiquitin-based probes, and diubiquitin. Angew. Chem. Int. Edn. Engl. 49, 10149–10153 (2010).
Hong, V., Presolski, S.I., Ma, C. & Finn, M.G. Analysis and optimization of copper-catalyzed azide-alkyne cycloaddition for bioconjugation. Angew. Chem. Int. Edn. Engl. 48, 9879–9883 (2009).
Plechanovová, A., Jaffray, E.G., Tatham, M.H., Naismith, J.H. & Hay, R.T. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489, 115–120 (2012).
Sommer, S., Weikart, N.D., Linne, U. & Mootz, H.D. Covalent inhibition of SUMO and ubiquitin-specific cysteine proteases by an in situ thiol-alkyne addition. Bioorg. Med. Chem. 21, 2511–2517 (2013).
Yang, B. et al. Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–906 (2012).
Narendra, D.P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate parkin. PLoS Biol. 8, e1000298 (2010).
Acknowledgements
We are grateful to the Medical Research Council Protein Phosphorylation and Ubiquitylation Unit Proteomics and DNA Sequencing Facilities. We are also grateful to I. Gilbert for support with chemistry instrumentation and access to modeling software. We also thank J.W. Chin (Medical Research Council Laboratory of Molecular Biology) for critical reading of the manuscript. This work was funded by the Scottish Funding Council, the UK Medical Research Council (MC_UU_12016/8) and pharmaceutical companies supporting the Division of Signal Transduction Therapy (AstraZeneca, Boehringer-Ingelheim, GlaxoSmithKline, Merck KGaA, Janssen Pharmaceutica and Pfizer). K.B. is funded by an A.J. Macdonald Menzies Charitable Trust Prize Studentship. M.M.K.M. is funded by a Wellcome Trust Senior Research Fellowship in Clinical Science (101022/Z/13/Z).
Author information
Authors and Affiliations
Contributions
S.V. conceived the project. Experiments were designed by S.V., M.M.K.M., K.-C.P. and M.S. K.-C.P. carried out experiments with assistance from M.S., C.H., Y.-C.L, P.M., K.B. and S.V. M.S. also carried out small-molecule synthesis. N.T.W. performed cloning. J.-C.C. and O.C. generated and provided fibroblasts derived from patients with PD. S.V. and M.M.K.M. wrote the manuscript with input from other authors.
Corresponding author
Ethics declarations
Competing interests
S.V., K.-C.P. and M.S. are authors on a patent that has been filed (PCT/GB2015/052860) relating to work presented in this article.
Supplementary information
Supplementary Text and Figures
Supplementary Results and Supplementary Figures 1–22. (PDF 10033 kb)
Rights and permissions
About this article
Cite this article
Pao, KC., Stanley, M., Han, C. et al. Probes of ubiquitin E3 ligases enable systematic dissection of parkin activation. Nat Chem Biol 12, 324–331 (2016). https://doi.org/10.1038/nchembio.2045
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.2045
This article is cited by
-
An expanded lexicon for the ubiquitin code
Nature Reviews Molecular Cell Biology (2023)
-
Spatiotemporal-resolved protein networks profiling with photoactivation dependent proximity labeling
Nature Communications (2022)
-
A new dawn beyond lysine ubiquitination
Nature Chemical Biology (2022)
-
Tools for the discovery of biopolymer producing cysteine relays
Biophysical Reviews (2021)
-
PINK1/PARKIN signalling in neurodegeneration and neuroinflammation
Acta Neuropathologica Communications (2020)