Abstract
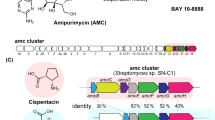
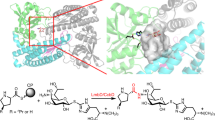
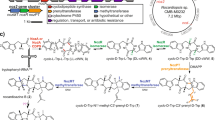
Cyclodipeptides and their derivatives belong to the diketopiperazine (DKP) family, which is comprised of a broad array of natural products that exhibit useful biological properties. In the few known DKP biosynthetic pathways, nonribosomal peptide synthetases (NRPSs) are involved in the synthesis of cyclodipeptides that constitute the DKP scaffold, except in the albonoursin (1) pathway. Albonoursin, or cyclo(α,β-dehydroPhe-α,β-dehydroLeu), is an antibacterial DKP produced by Streptomyces noursei. In this pathway, the formation of the cyclo(Phe-Leu) (2) intermediate is catalyzed by AlbC, a small protein unrelated to NRPSs. We demonstrated that AlbC uses aminoacyl-tRNAs as substrates to catalyze the formation of the DKP peptide bonds. Moreover, several other bacterial proteins, presenting moderate similarity to AlbC, also use aminoacyl-tRNAs to synthesize various cyclodipeptides. Therefore, AlbC and these related proteins belong to a newly defined family of enzymes that we have named cyclodipeptide synthases (CDPSs).
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ström, K., Sjogren, J., Broberg, A. & Schnurer, J. Lactobacillus plantarum MiLAB 393 produces the antifungal cyclic dipeptides cyclo(L-Phe-L-Pro) and cyclo(L-Phe-trans-4-OH-L-Pro) and 3-phenyllactic acid. Appl. Environ. Microbiol. 68, 4322–4327 (2002).
Kohn, H. & Widger, W. The molecular basis for the mode of action of bicyclomycin. Curr. Drug Targets Infect. Disord. 5, 273–295 (2005).
Kanoh, K. et al. Antitumor activity of phenylahistin in vitro and in vivo. Biosci. Biotechnol. Biochem. 63, 1130–1133 (1999).
Williams, D.E. et al. Ambewelamides A and B, antineoplastic epidithiapiperazinediones isolated from the lichen Usnea sp. Tetrahedr. Lett. 39, 9579–9582 (1998).
Waring, P. & Beaver, J. Gliotoxin and related epipolythiodioxopiperazines. Gen. Pharmacol. 27, 1311–1316 (1996).
Maiya, S., Grundmann, A., Li, S.M. & Turner, G. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem 7, 1062–1069 (2006).
Walzel, B., Riederer, B. & Keller, U. Mechanism of alkaloid cyclopeptide synthesis in the ergot fungus Claviceps purpurea. Chem. Biol. 4, 223–230 (1997).
Healy, F.G., Wach, M., Krasnoff, S.B., Gibson, D.M. & Loria, R. The txtAB genes of the plant pathogen Streptomyces acidiscabies encode a peptide synthetase required for phytotoxin thaxtomin A production and pathogenicity. Mol. Microbiol. 38, 794–804 (2000).
Schultz, A.W. et al. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J. Am. Chem. Soc. 130, 4507–4516 (2008).
Balibar, C.J. & Walsh, C.T. GliP, a multimodular nonribosomal peptide synthetase in Aspergillus fumigatus, makes the diketopiperazine scaffold of gliotoxin. Biochemistry 45, 15029–15038 (2006).
Gardiner, D.M., Cozijnsen, A.J., Wilson, L.M., Pedras, M.S. & Howlett, B.J. The sirodesmin biosynthetic gene cluster of the plant pathogenic fungus Leptosphaeria maculans. Mol. Microbiol. 53, 1307–1318 (2004).
Schwarzer, D., Mootz, H.D. & Marahiel, M.A. Exploring the impact of different thioesterase domains for the design of hybrid peptide synthetases. Chem. Biol. 8, 997–1010 (2001).
Gruenewald, S., Mootz, H.D., Stehmeier, P. & Stachelhaus, T. In vivo production of artificial nonribosomal peptide products in the heterologous host Escherichia coli. Appl. Environ. Microbiol. 70, 3282–3291 (2004).
Lautru, S., Gondry, M., Genet, R. & Pernodet, J.L. The albonoursin gene cluster of S. noursei: biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chem. Biol. 9, 1355–1364 (2002).
Fukushima, K., Yazawa, K. & Arai, T. Biological activities of albonoursin. J. Antibiot. (Tokyo) 26, 175–176 (1973).
Watanabe, K. et al. Protein-based peptide-bond formation by aminoacyl-tRNA protein transferase. Nature 449, 867–871 (2007).
Mainardi, J.L., Villet, R., Bugg, T.D., Mayer, C. & Arthur, M. Evolution of peptidoglycan biosynthesis under the selective pressure of antibiotics in Gram-positive bacteria. FEMS Microbiol. Rev. 32, 386–408 (2008).
Rost, B. Twilight zone of protein sequence alignments. Protein Eng. 12, 85–94 (1999).
Combet, C., Blanchet, C., Geourjon, C. & Deleage, G. NPS@: network protein sequence analysis. Trends Biochem. Sci. 25, 147–150 (2000).
Loria, R. et al. Thaxtomin biosynthesis: the path to plant pathogenicity in the genus Streptomyces. Antonie Van Leeuwenhoek 94, 3–10 (2008).
Gondry, M. et al. Cyclic dipeptide oxidase from Streptomyces noursei. Isolation, purification and partial characterization of a novel, amino acyl alpha,beta-dehydrogenase. Eur. J. Biochem. 268, 1712–1721 (2001).
McLean, K.J. et al. Characterization of active site structure in CYP121: a cytochrome P450 essential for viability of Mycobacterium tuberculosis H37Rv. J. Biol. Chem. 283, 33406–33416 (2008).
Belin, P. et al. Identification and structural basis of the reaction catalyzed by CYP121, an essential cytochrome P450 in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA (in the press).
Tang, M.R., Sternberg, D., Behr, R.K., Sloma, A. & Berka, R.M. Use of transcriptional profiling & bioinformatics to solve production problems: eliminating red pigment production in a Bacillus subtilis strain producing hyaluronic acid. Ind. Biotechnol. 2, 66–74 (2006).
Uffen, R.L. & Canale-Parola, E. Synthesis of pulcherriminic acid by Bacillus subtilis. J. Bacteriol. 111, 86–93 (1972).
RajBhandary, U.L. & Soll, D. Aminoacyl-tRNAs, the bacterial cell envelope, and antibiotics. Proc. Natl. Acad. Sci. USA 105, 5285–5286 (2008).
Abramochkin, G. & Shrader, T.E. Aminoacyl-tRNA recognition by the leucyl/phenylalanyl-tRNA-protein transferase. J. Biol. Chem. 271, 22901–22907 (1996).
Villet, R. et al. Idiosyncratic features in tRNAs participating in bacterial cell wall synthesis. Nucleic Acids Res. 35, 6870–6883 (2007).
Kanzaki, H., Imura, D., Nitoda, T. & Kawazu, K. Enzymatic dehygrogenation of cyclo(L-Leu-L-Phe) to a bioactive derivative, albonoursin. J. Mol. Catal. B Enzym. 6, 265–270 (1999).
Abramochkin, G. & Shrader, T.E. The leucyl/phenylalanyl-tRNA-protein transferase. Overexpression and characterization of substrate recognition, domain structure, and secondary structure. J. Biol. Chem. 270, 20621–20628 (1995).
Chen, Y.-H., Liou, S.-E. & Chen, C.-C. Two-step mass spectrometric approach for the identification of diketopiperazines in chicken essence. Eur. Food Res. Technol. 218, 589–597 (2004).
Stark, T. & Hofmann, T. Structures, sensory activity, and dose/response functions of 2,5-diketopiperazines in roasted cocoa nibs (Theobroma cacao). J. Agric. Food Chem. 53, 7222–7231 (2005).
Falick, A.M., Hines, W.M., Medzihradszky, K.F., Baldwin, M.A. & Gibson, B.W. Low-mass ions produced from peptides by high-energy collision-induced dissociation in tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 4, 882–893 (1993).
Papayannopoulos, I.A. The interpretation of collision-induced dissociation tandem mass spectra of peptides. Mass Spectrom. Rev. 14, 49–73 (1995).
Roepstorff, P. & Fohlman, J. Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601 (1984).
Johnson, R.S., Martin, S.A., Biemann, K., Stults, J.T. & Watson, J.T. Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass spectrometer: differentiation of leucine and isoleucine. Anal. Chem. 59, 2621–2625 (1987).
Jeedigunta, S., Krenisky, J.M. & Kerr, R.G. Diketopiperazines as advanced intermediates in the biosynthesis of Ecteinascidins. Tetrahedron 56, 3303–3307 (2000).
Altschul, S.F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Corpet, F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 (1988).
Acknowledgements
We thank C. Olaizola and M. Bahut for technical assistance, S. Zinn-Justin and J. Baillon for helpful discussion, and M. Moutiez for the critical reading of the manuscript. We are indebted to M. Springer (Institut de Biologie Physico-Chimique) for the plasmid pBI and to F. Doucet-Populaire (Université Paris Descartes), T. Msadek (Pasteur Institute), A. Bolotin (Institut National de la Recherche Agronomique), T. Baba (Juntendo University) and E. Krin (Pasteur Institute) for the gifts of bacterial strains and bacterial genomic DNAs. This study was supported by Commissariat à l'Energie Atomique, Centre National de la Recherche Scientifique, Université Paris-Sud 11, Pôle de Recherche et d'Enseignement Supérieur UniverSud Paris and Kyowa Hakko Bio Co. Ltd. L.S. is a recipient of a doctoral fellowship from Commissariat à l'Energie Atomique.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
M.G., L.S., P.B., R.T., S.-i.H., R.G. and J.-L.P. are inventors in patent applications related to the use of CDPSs for dipeptide synthesis. The research performed was supported in part by Kyowa Hakko Bio Co. Ltd, a company that is a co-owner of the patent applications. S.-i.H. is an employee of Kyowa Hakko Bio Co. Ltd.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Table 1 and Supplementary Methods (PDF 1459 kb)
Rights and permissions
About this article
Cite this article
Gondry, M., Sauguet, L., Belin, P. et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond–forming enzymes. Nat Chem Biol 5, 414–420 (2009). https://doi.org/10.1038/nchembio.175
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.175
This article is cited by
-
Unveiling an indole alkaloid diketopiperazine biosynthetic pathway that features a unique stereoisomerase and multifunctional methyltransferase
Nature Communications (2023)
-
Biological and Sensing Applications of a Few 1,3,4-Oxadiazole Based Donor-Acceptor Systems
Journal of Fluorescence (2023)
-
Genome mining, antimicrobial and plant growth-promoting potentials of halotolerant Bacillus paralicheniformis ES-1 isolated from salt mine
Molecular Genetics and Genomics (2023)
-
Active site remodelling of a cyclodipeptide synthase redefines substrate scope
Communications Chemistry (2022)
-
Mating type specific transcriptomic response to sex inducing pheromone in the pennate diatom Seminavis robusta
The ISME Journal (2021)