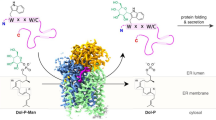
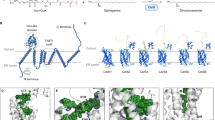
Abstract
Secondary structure refolding is a key event in biology as it modulates the conformation of many proteins in the cell, generating functional or aberrant states. The crystal structures of mannosyltransferase PimA reveal an exceptional flexibility of the protein along the catalytic cycle, including β-strand–to–α-helix and α-helix–to–β-strand transitions. These structural changes modulate catalysis and are promoted by interactions of the protein with anionic phospholipids in the membrane.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Korduláková, J. et al. J. Biol. Chem. 277, 31335–31344 (2002).
Guerin, M.E. et al. J. Biol. Chem. 284, 25687–25696 (2009).
Guerin, M.E., Kordulakova, J., Alzari, P.M., Brennan, P.J. & Jackson, M. J. Biol. Chem. 285, 33577–33583 (2010).
Varki, A. et al. Essentials of Glycobiology (Cold Spring Harbor Laboratory Press, New York, 2009).
Lairson, L.L., Henrissat, B., Davies, G.J. & Withers, S.G. Annu. Rev. Biochem. 77, 521–555 (2008).
Albesa-Jové, D., Giganti, D., Jackson, M., Alzari, P.M. & Guerin, M.E. Glycobiology 24, 108–124 (2014).
Guerin, M.E. et al. J. Biol. Chem. 282, 20705–20714 (2007).
Guerin, M.E. et al. J. Biol. Chem. 284, 21613–21625 (2009).
Giganti, D. et al. J. Biol. Chem. 288, 29797–29808 (2013).
Hu, Y. et al. Proc. Natl. Acad. Sci. USA 100, 845–849 (2003).
Ge, C., Georgiev, A., Öhman, A., Wieslander, Å. & Kelly, A.A. J. Biol. Chem. 286, 6669–6684 (2011).
Schmidt, H. et al. Proc. Natl. Acad. Sci. USA 109, 6253–6258 (2012).
Rebecchi, M.J., Eberhardt, R., Delaney, T., Ali, S. & Bittman, R. J. Biol. Chem. 268, 1735–1741 (1993).
Lindeberg, M.I., Zakharov, S.D. & Cramer, W.A.J. Mol. Biol. 295, 679–692 (2000).
Mosbahi, K. et al. J. Biol. Chem. 279, 22145–22151 (2004).
Tilley, S.J., Orlova, E.V., Gilbert, R.J., Andrew, P.W. & Saibil, H.R. Cell 121, 247–256 (2005).
le Maire, A. et al. Nat. Struct. Mol. Biol. 17, 801–807 (2010).
Burmann, B.M. et al. Cell 150, 291–303 (2012).
Lucast, L.J., Batey, R.T. & Doudna, J.A. Biotechniques 30, 544–546 (2001).
Kabsch, W. J. Appl. Crystallogr. 26, 795–800 (1993).
McCoy, A.J. et al. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P. & Cowtan, K. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Blanc, E. et al. Acta Crystallogr. D Biol. Crystallogr. 60, 2210–2221 (2004).
Schaeffer, F. et al. Biochemistry 41, 2106–2114 (2002).
Wiseman, T., Williston, S., Brandts, J.F. & Lin, L.N. Anal. Biochem. 179, 131–137 (1989).
Fiske, C.H. & Subbarow, Y. J. Biol. Chem. 66, 375–400 (1925).
Montagner, C. et al. Biochemistry 46, 1878–1887 (2007).
Wriggers, W. Biophys. Rev. 2, 21–27 (2010).
Svergun, D.I., Barberato, C. & Koch, M.H.J. J. Appl. Crystallogr. 28, 768–773 (1995).
Acknowledgements
We acknowledge A. Haouz and P. Weber (Institut Pasteur, France) for help with robotic crystallization; S. Lopez Fernandez and P. Arrasate (Structural Glycobiology Group, Unit of Biophysics, Spain) for technical assistance; and the European Synchrotron Radiation Facility (ESRF), the French National Synchrotron SOLEIL, the Diamond Light Source (DLS) and the Swiss Light Source (SLS) for granting access to synchrotron radiation facilities and their staff for the onsite assistance. We specially thank the BioStruct-X project to support access to structural biology facilities. Technical support provided by Universidad del País Vasco/Euskal Herriko Unibertsitatea and Ministerio de Ciencia e Innovación is acknowledged. We also thank all members of the Structural Glycobiology Group for valuable scientific discussions. This work was supported by the European Community's Sixth and Seventh Framework Programmes (contracts LSHP-CT-2005-018923 and HEALTH-F3-2011-260872), the Institut Pasteur, the Spanish Ministry of Science and Innovation (contracts SAF2010-19096 and BIO2013-49022-C2-2-R), IKERBASQUE, the Basque Foundation for Science, the Basque Government and the Fundación Biofísica Bizkaia.
Author information
Authors and Affiliations
Contributions
M.E.G. and P.M.A. conceived the project. D.G., D.A.-J. and M.B. performed the crystallographic studies. S.U., A.R.-U., M.A.M., N.B. and N.C. prepared and characterized wild-type PimA and the different mutants. D.G., M.A.M., M.B., A.C., M.E.G. & P.M.A. analyzed the results. D.G., A.C., M.E.G. & P.M.A. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–15 and Supplementary Table 1. (PDF 2845 kb)
Supplementary Video 1
Structural transition between the extended and compact conformational states of apo PimA (MPG 522 kb)
Supplementary Video 2
Structural transition between apo PimA and GDP-bound PimA involving the reshuffling of secondary structural elements from the N-terminal domain. (MPG 1520 kb)
Rights and permissions
About this article
Cite this article
Giganti, D., Albesa-Jové, D., Urresti, S. et al. Secondary structure reshuffling modulates glycosyltransferase function at the membrane. Nat Chem Biol 11, 16–18 (2015). https://doi.org/10.1038/nchembio.1694
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1694
This article is cited by
-
Self-recycling and partially conservative replication of mycobacterial methylmannose polysaccharides
Communications Biology (2023)
-
Secondary-structure switch regulates the substrate binding of a YopJ family acetyltransferase
Nature Communications (2021)
-
Jawsamycin exhibits in vivo antifungal properties by inhibiting Spt14/Gpi3-mediated biosynthesis of glycosylphosphatidylinositol
Nature Communications (2020)
-
Crystal structure of lipid A disaccharide synthase LpxB from Escherichia coli
Nature Communications (2018)
-
Structural basis for selective recognition of acyl chains by the membrane-associated acyltransferase PatA
Nature Communications (2016)