Abstract
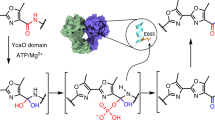
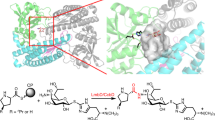
Despite intensive research, the cyclodehydratase responsible for azoline biogenesis in thiazole/oxazole-modified microcin (TOMM) natural products remains enigmatic. The collaboration of two proteins, C and D, is required for cyclodehydration. The C protein is homologous to E1 ubiquitin-activating enzymes, whereas the D protein is within the YcaO superfamily. Recent studies have demonstrated that TOMM YcaOs phosphorylate amide carbonyl oxygens to facilitate azoline formation. Here we report the X-ray crystal structure of an uncharacterized YcaO from Escherichia coli (Ec-YcaO). Ec-YcaO harbors an unprecedented fold and ATP-binding motif. This motif is conserved among TOMM YcaOs and is required for cyclodehydration. Furthermore, we demonstrate that the C protein regulates substrate binding and catalysis and that the proline-rich C terminus of the D protein is involved in C protein recognition and catalysis. This study identifies the YcaO active site and paves the way for the characterization of the numerous YcaO domains not associated with TOMM biosynthesis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Strader, M.B. et al. A proteomic and transcriptomic approach reveals new insight into β-methylthiolation of Escherichia coli ribosomal protein S12. Mol. Cell. Proteomics 10, M110005199 (2011).
Tenorio, E. et al. Systematic characterization of Escherichia coli genes/ORFs affecting biofilm formation. FEMS Microbiol. Lett. 225, 107–114 (2003).
Melby, J.O., Nard, N.J. & Mitchell, D.A. Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr. Opin. Chem. Biol. 15, 369–378 (2011).
Arnison, P.G. et al. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30, 108–160 (2013).
Molloy, E.M., Cotter, P.D., Hill, C., Mitchell, D.A. & Ross, R.P. Streptolysin S-like virulence factors: the continuing sagA. Nat. Rev. Microbiol. 9, 670–681 (2011).
Li, Y.M., Milne, J.C., Madison, L.L., Kolter, R. & Walsh, C.T. From peptide precursors to oxazole and thiazole-containing peptide antibiotics: microcin B17 synthase. Science 274, 1188–1193 (1996).
Dunbar, K.L., Melby, J.O. & Mitchell, D.A. YcaO domains use ATP to activate amide backbones during peptide cyclodehydrations. Nat. Chem. Biol. 8, 569–575 (2012).
Koehnke, J. et al. The cyanobactin heterocyclase enzyme: a processive adenylase that operates with a defined order of reaction. Angew. Chem. Int. Ed. Engl. 52, 13991–13996 (2013).
Lee, S.W. et al. Discovery of a widely distributed toxin biosynthetic gene cluster. Proc. Natl. Acad. Sci. USA 105, 5879–5884 (2008).
Schmidt, E.W. et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc. Natl. Acad. Sci. USA 102, 7315–7320 (2005).
McIntosh, J.A. & Schmidt, E.W. Marine molecular machines: heterocyclization in cyanobactin biosynthesis. ChemBioChem 11, 1413–1421 (2010).
Milne, J.C. et al. Cofactor requirements and reconstitution of microcin B17 synthetase: a multienzyme complex that catalyzes the formation of oxazoles and thiazoles in the antibiotic microcin B17. Biochemistry 38, 4768–4781 (1999).
Milne, J.C., Eliot, A.C., Kelleher, N.L. & Walsh, C.T. ATP/GTP hydrolysis is required for oxazole and thiazole biosynthesis in the peptide antibiotic microcin B17. Biochemistry 37, 13250–13261 (1998).
Schulman, B.A. & Harper, J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 10, 319–331 (2009).
Mitchell, D.A. et al. Structural and functional dissection of the heterocyclic peptide cytotoxin streptolysin S. J. Biol. Chem. 284, 13004–13012 (2009).
Dunbar, K.L. & Mitchell, D.A. Insights into the mechanism of peptide cyclodehydrations achieved through the chemoenzymatic generation of amide derivatives. J. Am. Chem. Soc. 135, 8692–8701 (2013).
Huo, L., Rachid, S., Stadler, M., Wenzel, S.C. & Muller, R. Synthetic biotechnology to study and engineer ribosomal bottromycin biosynthesis. Chem. Biol. 19, 1278–1287 (2012).
Hou, Y. et al. Structure and biosynthesis of the antibiotic bottromycin D. Org. Lett. 14, 5050–5053 (2012).
Gomez-Escribano, J.P., Song, L., Bibb, M.J. & Challis, G.L. Posttranslational β-methylation and macrolactamidination in the biosynthesis of the bottromycin complex of ribosomal peptide antibiotics. Chem. Sci. 3, 3522–3525 (2012).
Crone, W.J.K., Leeper, F.J. & Truman, A.W. Identification and characterization of the gene cluster for the anti-MRSA antibiotic bottromycin: expanding the biosynthetic diverstiy of ribosomal peptides. Chem. Sci. 3, 3516–3521 (2012).
Breil, B.T., Ludden, P.W. & Triplett, E.W. DNA sequence and mutational analysis of genes involved in the production and resistance of the antibiotic peptide trifolitoxin. J. Bacteriol. 175, 3693–3702 (1993).
Webb, M.R. A continuous spectrophotometric assay for inorganic phosphate and for measuring phosphate release kinetics in biological systems. Proc. Natl. Acad. Sci. USA 89, 4884–4887 (1992).
Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 (2010).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003).
Hunter, S. et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 40, D306–D312 (2012).
Breil, B., Borneman, J. & Triplett, E.W. A newly discovered gene, tfuA, involved in the production of the ribosomally synthesized peptide antibiotic trifolitoxin. J. Bacteriol. 178, 4150–4156 (1996).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Oman, T.J. & van der Donk, W.A. Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat. Chem. Biol. 6, 9–18 (2010).
Melby, J.O., Dunbar, K.L., Trinh, N.Q. & Mitchell, D.A. Selectivity, directionality, and promiscuity in peptide processing from a Bacillus sp. Al Hakam cyclodehydratase. J. Am. Chem. Soc. 134, 5309–5316 (2012).
Denessiouk, K.A. & Johnson, M.S. When fold is not important: a common structural framework for adenine and AMP binding in 12 unrelated protein families. Proteins 38, 310–326 (2000).
Zhang, Y., Morar, M. & Ealick, S.E. Structural biology of the purine biosynthetic pathway. Cell. Mol. Life Sci. 65, 3699–3724 (2008).
Fawaz, M.V., Topper, M.E. & Firestine, S.M. The ATP-grasp enzymes. Bioorg. Chem. 39, 185–191 (2011).
Rakus, J.F. et al. Evolution of enzymatic activities in the enolase superfamily: d-mannonate dehydratase from Novosphingobium aromaticivorans. Biochemistry 46, 12896–12908 (2007).
Chen, W., Biswas, T., Porter, V.R., Tsodikov, O.V. & Garneau-Tsodikova, S. Unusual regioversatility of acetyltransferase Eis, a cause of drug resistance in XDR-TB. Proc. Natl. Acad. Sci. USA 108, 9804–9808 (2011).
Selvy, P.E., Lavieri, R.R., Lindsley, C.W. & Brown, H.A. Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 (2011).
Bhatnagar, R.S., Futterer, K., Waksman, G. & Gordon, J.I. The structure of myristoyl-CoA:protein N-myristoyltransferase. Biochim. Biophys. Acta 1441, 162–172 (1999).
Climie, S.C., Carreras, C.W. & Santi, D.V. Complete replacement set of amino acids at the C terminus of thymidylate synthase: quantitative structure-activity relationship of mutants of an enzyme. Biochemistry 31, 6032–6038 (1992).
Regni, C.A. et al. How the MccB bacterial ancestor of ubiquitin E1 initiates biosynthesis of the microcin C7 antibiotic. EMBO J. 28, 1953–1964 (2009).
Donia, M.S. et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat. Chem. Biol. 2, 729–735 (2006).
Izawa, M., Kawasaki, T. & Hayakawa, Y. Cloning and heterologous expression of the thioviridamide biosynthesis gene cluster from Streptomyces olivoviridis. Appl. Environ. Microbiol. 79, 7110–7113 (2013).
Sievers, F. et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539 (2011).
Atkinson, H.J., Morris, J.H., Ferrin, T.E. & Babbitt, P.C. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE 4, e4345 (2009).
Tamura, K. et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Larkin, M.A. et al. Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 (2007).
Laskowski, R.A. & Swindells, M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 51, 2778–2786 (2011).
Ashkenazy, H., Erez, E., Martz, E., Pupko, T. & Ben-Tal, N. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 (2010).
Bond, C.S. & Schuttelkopf, A.W. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. D Biol. Crystallogr. 65, 510–512 (2009).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr. 67, 293–302 (2011).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 (2008).
Emsley, P., Lohkamp, B., Scott, W.G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Vagin, A.A. et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 60, 2184–2195 (2004).
Laskowski, R.A., MacArthur, M.W., Moss, D.S. & Thornton, J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Acknowledgements
We are grateful to C. Deane and K. Taylor for the generation of select BalhD mutants and J. Melby for assistance with collecting MS/MS data. This work was supported by the US National Institutes of Health (NIH) (1R01 GM097142 to D.A.M., 1R01 GM102602 to S.K.N. and 2T32 GM070421 to K.L.D., B.J.B. and J.R.C.). Additional support was from the Harold R. Snyder Fellowship (University of Illinois at Urbana-Champaign (UIUC) Department of Chemistry to K.L.D.), the Robert C. and Carolyn J. Springborn Endowment (UIUC Department of Chemistry to B.J.B.), the National Science Foundation Graduate Research Fellowship (DGE-1144245 to B.J.B.) and the University of Illinois Distinguished Fellowship (UIUC Graduate College to J.R.C.) The Bruker UltrafleXtreme MALDI TOF/TOF mass spectrometer was purchased in part with a grant from the NIH–National Center for Research Resources (S10 RR027109 A).
Author information
Authors and Affiliations
Contributions
Experiments were designed by D.A.M., S.K.N., K.L.D., J.R.C., C.L.C. and B.J.B. and were performed by K.L.D., J.R.C., C.L.C. and B.J.B. The manuscript was written by D.A.M., K.L.D. and J.R.C. with critical editorial input from S.K.N., C.L.C. and B.J.B. The overall study was conceived and managed by D.A.M. with S.K.N. overseeing all aspects of protein structure determination.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–3 and Supplementary Figures 1–27. (PDF 16297 kb)
Rights and permissions
About this article
Cite this article
Dunbar, K., Chekan, J., Cox, C. et al. Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis. Nat Chem Biol 10, 823–829 (2014). https://doi.org/10.1038/nchembio.1608
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1608
This article is cited by
-
Functional elucidation of TfuA in peptide backbone thioamidation
Nature Chemical Biology (2021)
-
Minimal lactazole scaffold for in vitro thiopeptide bioengineering
Nature Communications (2020)
-
Natural thiopeptides as a privileged scaffold for drug discovery and therapeutic development
Medicinal Chemistry Research (2019)
-
Klebsazolicin inhibits 70S ribosome by obstructing the peptide exit tunnel
Nature Chemical Biology (2017)
-
Dissection of goadsporin biosynthesis by in vitro reconstitution leading to designer analogues expressed in vivo
Nature Communications (2017)