Abstract
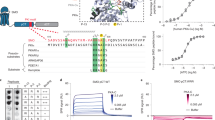
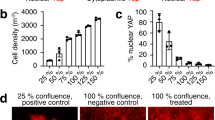
Hedgehog (Hh) signaling determines cell fate during development and can drive tumorigenesis. We performed a screen for new compounds that can impinge on Hh signaling downstream of Smoothened (Smo). A series of cyclohexyl-methyl aminopyrimidine chemotype compounds ('CMAPs') were identified that could block pathway signaling in a Smo-independent manner. In addition to inhibiting Hh signaling, the compounds generated inositol phosphates through an unknown GPCR. Correlation of GPCR mRNA expression levels with compound activity across cell lines suggested the target to be the orphan receptor GPR39. RNA interference or cDNA overexpression of GPR39 demonstrated that the receptor is necessary for compound activity. We propose a model in which CMAPs activate GPR39, which signals to the Gli transcription factors and blocks signaling. In addition to the discovery of GPR39 as a new target that impinges on Hh signaling, we report on small-molecule modulators of the receptor that will enable in vitro interrogation of GPR39 signaling in different cellular contexts.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ingham, P.W., Nakano, Y. & Seger, C. Mechanisms and functions of Hedgehog signalling across the metazoa. Nat. Rev. Genet. 12, 393–406 (2011).
Jiang, J. & Hui, C.C. Hedgehog signaling in development and cancer. Dev. Cell 15, 801–812 (2008).
Rubin, L.L. & de Sauvage, F.J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 5, 1026–1033 (2006).
Hahn, H. et al. Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85, 841–851 (1996).
Xie, J. et al. Activating Smoothened mutations in sporadic basal-cell carcinoma. Nature 391, 90–92 (1998).
Taylor, M.D. et al. Mutations in SUFU predispose to medulloblastoma. Nat. Genet. 31, 306–310 (2002).
Northcott, P.A. et al. Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma. Nat. Genet. 41, 465–472 (2009).
Pastorino, L. et al. Identification of a SUFU germline mutation in a family with Gorlin syndrome. Am. J. Med. Genet. A 149A, 1539–1543 (2009).
Northcott, P.A. et al. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature 488, 49–56 (2012).
Ragazzini, P. et al. Amplification of CDK4, MDM2, SAS and GLI genes in leiomyosarcoma, alveolar and embryonal rhabdomyosarcoma. Histol. Histopathol. 19, 401–411 (2004).
Kinzler, K.W. et al. Identification of an amplified, highly expressed gene in a human glioma. Science 236, 70–73 (1987).
Skvara, H. et al. Topical treatment of basal cell carcinomas in nevoid basal cell carcinoma syndrome with a smoothened inhibitor. J. Invest. Dermatol. 131, 1735–1744 (2011).
Low, J.A. & de Sauvage, F.J. Clinical experience with Hedgehog pathway inhibitors. J. Clin. Oncol. 28, 5321–5326 (2010).
Buonamici, S. et al. Interfering with resistance to smoothened antagonists by inhibition of the pi3k pathway in medulloblastoma. Sci. Transl. Med. 2, 1–8 (2010).
Yauch, R.L. et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science 326, 572–574 (2009).
Metcalfe, C. & de Sauvage, F.J. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 71, 5057–5061 (2011).
Dijkgraaf, G.J. et al. Small molecule inhibition of GDC-0449 refractory smoothened mutants and downstream mechanisms of drug resistance. Cancer Res. 71, 435–444 (2011).
Miller-Moslin, K. et al. 1-Amino-4-benzylphthalazines as orally bioavailable smoothened antagonists with antitumor activity. J. Med. Chem. 52, 3954–3968 (2009).
Frank-Kamenetsky, M. et al. Small-molecule modulators of Hedgehog signaling: identification and characterization of Smoothened agonists and antagonists. J. Biol. 1, 10 (2002).
Chen, J.K., Taipale, J., Cooper, M.K. & Beachy, P.A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 16, 2743–2748 (2002).
Romer, J.T. et al. Suppression of the Shh pathway using a small molecule inhibitor eliminates medulloblastoma in Ptc1+/− p53−/− mice. Cancer Cell 6, 229–240 (2004).
Peukert, S. et al. Discovery of NVP-LEQ506, a second-generation inhibitor of Smoothened. ChemMedChem 8, 1261–1265 (2013).
Nachtergaele, S. et al. Oxysterols are allosteric activators of the oncoprotein Smoothened. Nat. Chem. Biol. 8, 211–220 (2012).
Huang, S.M. et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature 461, 614–620 (2009).
Nidhi Glick, M., Davies, J.W. & Jenkins, J.L. Prediction of biological targets for compounds using multiple-category Bayesian models trained on chemogenomics databases. J. Chem. Inf. Model. 46, 1124–1133 (2006).
Waschek, J.A., Dicicco-Bloom, E., Nicot, A. & Lelievre, V. Hedgehog signaling: new targets for GPCRs coupled to cAMP and protein kinase A. Ann. NY Acad. Sci. 1070, 120–128 (2006).
Lelievre, V. et al. Disruption of the PACAP gene promotes medulloblastoma in ptc1 mutant mice. Dev. Biol. 313, 359–370 (2008).
Newman-Tancredi, A., Cussac, D., Marini, L. & Millan, M.J. Antibody capture assay reveals bell-shaped concentration-response isotherms for h5-HT1A receptor-mediated Gαi3 activation: conformational selection by high-efficacy agonists, and relationship to trafficking of receptor signaling. Mol. Pharmacol. 62, 590–601 (2002).
Depoortere, I. GI functions of GPR39: novel biology. Curr. Opin. Pharmacol. 12, 647–652 (2012).
Popovics, P. & Stewart, A.J. GPR39: a Zn2+-activated G protein–coupled receptor that regulates pancreatic, gastrointestinal and neuronal functions. Cell. Mol. Life Sci. 68, 85–95 (2011).
Moechars, D. et al. Altered gastrointestinal and metabolic function in the GPR39-obestatin receptor-knockout mouse. Gastroenterology 131, 1131–1141 (2006).
Egerod, K.L. et al. β-cell specific overexpression of GPR39 protects against streptozotocin-induced hyperglycemia. Int. J. Endocrinol. 2011, 401258 (2011).
Holst, B. et al. G protein–coupled receptor 39 deficiency is associated with pancreatic islet dysfunction. Endocrinology 150, 2577–2585 (2009).
Petersen, P.S. et al. Deficiency of the GPR39 receptor is associated with obesity and altered adipocyte metabolism. FASEB J. 25, 3803–3814 (2011).
Tremblay, F. et al. Disruption of G protein–coupled receptor 39 impairs insulin secretion in vivo. Endocrinology 150, 2586–2595 (2009).
Verhulst, P.J. et al. GPR39, a receptor of the ghrelin receptor family, plays a role in the regulation of glucose homeostasis in a mouse model of early onset diet-induced obesity. J. Neuroendocrinol. 23, 490–500 (2011).
Zhang, J.V. et al. Obestatin, a peptide encoded by the ghrelin gene, opposes ghrelin's effects on food intake. Science 310, 996–999 (2005).
Holst, B. et al. GPR39 signaling is stimulated by zinc ions but not by obestatin. Endocrinology 148, 13–20 (2007).
Dong, X.Y. et al. Is GPR39 the natural receptor of obestatin? Peptides 30, 431–438 (2009).
Yasuda, S. et al. Isolation of Zn2+ as an endogenous agonist of GPR39 from fetal bovine serum. J. Recept. Signal Transduct. Res. 27, 235–246 (2007).
Markus, B. et al. Chemical probe identification platform for orphan GPCRs using focused compound screening: GPR39 as a case example. ACS Med. Chem. Lett. 4, 1079–1084 (2013).
Lauth, M., Bergstrom, A. & Toftgard, R. Phorbol esters inhibit the Hedgehog signalling pathway downstream of Suppressor of Fused, but upstream of Gli. Oncogene 26, 5163–5168 (2007).
Atwood, S.X., Li, M., Lee, A., Tang, J.Y. & Oro, A.E. GLI activation by atypical protein kinase Cι/λ regulates the growth of basal cell carcinomas. Nature 494, 484–488 (2013).
Axelrod, M. et al. Combinatorial drug screening identifies compensatory pathway interactions and adaptive resistance mechanisms. Oncotarget 4, 622–635 (2013).
Ji, Z., Mei, F.C., Xie, J. & Cheng, X. Oncogenic KRAS activates hedgehog signaling pathway in pancreatic cancer cells. J. Biol. Chem. 282, 14048–14055 (2007).
Fogarty, M.P., Emmenegger, B.A., Grasfeder, L.L., Oliver, T.G. & Wechsler-Reya, R.J. Fibroblast growth factor blocks Sonic hedgehog signaling in neuronal precursors and tumor cells. Proc. Natl. Acad. Sci. USA 104, 2973–2978 (2007).
Berridge, M.J., Downes, C.P. & Hanley, M.R. Lithium amplifies agonist-dependent phosphatidylinositol responses in brain and salivary glands. Biochem. J. 206, 587–595 (1982).
Seuwen, K., Lagarde, A. & Pouyssegur, J. Deregulation of hamster fibroblast proliferation by mutated ras oncogenes is not mediated by constitutive activation of phosphoinositide-specific phospholipase C. EMBO J. 7, 161–168 (1988).
Berridge, M.J. & Irvine, R.F. Inositol phosphates and cell signalling. Nature 341, 197–205 (1989).
Salomon, Y. Adenylate cyclase assay. Adv. Cycl. Nucleot. Res. 10, 35–55 (1979).
Acknowledgements
We thank A. Abrams for assistance with production of a figure for this publication.
Author information
Authors and Affiliations
Contributions
F.B., J.F.K. III, K.S., R.K.J. and S.J.L. designed experiments. A.C., J.A.D., A.B., A.R., S.J.L. and F.H. performed Hh pathway assays. F.B., B.G., S.V., V.B., B.W., A.S. and A.C. designed and performed GPCR-focused cell-based experiments. X.W. performed the initial screen. L.A.L., T.B.S., P.R. and R.K.J. synthesized compounds. S.P. and L.L. performed cell-free experiments. H.R., P.G., M.S., J.J. and T.B. designed and performed proteomics experiments. T.B., J.A.P., V.M., P.M.F. and J.A.T. provided intellectual input during the target identification campaign. S.J.L., R.K.J. and F.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors are employees of Novartis and Cellzome Ag. The research was fully funded by Novartis.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Figures 1–17, Supplementary Tables 1–3 and Supplementary Note. (PDF 33061 kb)
Rights and permissions
About this article
Cite this article
Bassilana, F., Carlson, A., DaSilva, J. et al. Target identification for a Hedgehog pathway inhibitor reveals the receptor GPR39. Nat Chem Biol 10, 343–349 (2014). https://doi.org/10.1038/nchembio.1481
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1481
This article is cited by
-
Targeted protein degradation reveals BET bromodomains as the cellular target of Hedgehog pathway inhibitor-1
Nature Communications (2023)
-
TC-G 1008 facilitates epileptogenesis by acting selectively at the GPR39 receptor but non-selectively activates CREB in the hippocampus of pentylenetetrazole-kindled mice
Cellular and Molecular Life Sciences (2023)
-
Discovery of new GPCR ligands to illuminate new biology
Nature Chemical Biology (2017)
-
An in vivo multiplexed small-molecule screening platform
Nature Methods (2016)
-
GPR39 marks specific cells within the sebaceous gland and contributes to skin wound healing
Scientific Reports (2015)