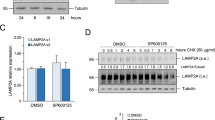
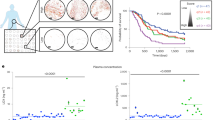
Abstract
Lysyl-tRNA synthetase (KRS), a protein synthesis enzyme in the cytosol, relocates to the plasma membrane after a laminin signal and stabilizes a 67-kDa laminin receptor (67LR) that is implicated in cancer metastasis; however, its potential as an antimetastatic therapeutic target has not been explored. We found that the small compound BC-K-YH16899, which binds KRS, impinged on the interaction of KRS with 67LR and suppressed metastasis in three different mouse models. The compound inhibited the KRS-67LR interaction in two ways. First, it directly blocked the association between KRS and 67LR. Second, it suppressed the dynamic movement of the N-terminal extension of KRS and reduced membrane localization of KRS. However, it did not affect the catalytic activity of KRS. Our results suggest that specific modulation of a cancer-related KRS-67LR interaction may offer a way to control metastasis while avoiding the toxicities associated with inhibition of the normal functions of KRS.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Steeg, P.S. Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 12, 895–904 (2006).
Berno, V. et al. The 67 kDa laminin receptor increases tumor aggressiveness by remodeling laminin-1. Endocr. Relat. Cancer 12, 393–406 (2005).
Givant-Horwitz, V., Davidson, B. & Reich, R. Laminin-induced signaling in tumor cells. Cancer Lett. 223, 1–10 (2005).
Kim, D.G. et al. Interaction of two translational components, lysyl-tRNA synthetase and p40/37LRP, in plasma membrane promotes laminin-dependent cell migration. FASEB J. 26, 4142–4159 (2012).
Scheiman, J., Tseng, J.C., Zheng, Y. & Meruelo, D. Multiple functions of the 37/67-kD laminin receptor make it a suitable target for novel cancer gene therapy. Mol. Ther. 18, 63–74 (2010).
van den Brûle, F.A. et al. Expression of the 67-kD laminin receptor, galectin-1, and galectin-3 in advanced human uterine adenocarcinoma. Hum. Pathol. 27, 1185–1191 (1996).
Sanjuán, X. et al. Overexpression of the 67-kD laminin receptor correlates with tumour progression in human colorectal carcinoma. J. Pathol. 179, 376–380 (1996).
Martignone, S. et al. Prognostic significance of the 67-kilodalton laminin receptor expression in human breast carcinomas. J. Natl. Cancer Inst. 85, 398–402 (1993).
Fontanini, G. et al. 67-Kilodalton laminin receptor expression correlates with worse prognostic indicators in non-small cell lung carcinomas. Clin. Cancer Res. 3, 227–231 (1997).
Li, D. et al. 67-kDa laminin receptor in human bile duct carcinoma. Eur. Surg. Res. 42, 168–173 (2009).
Batissoco, A.C., Auricchio, M.T., Kimura, L., Tabith-Junior, A. & Mingroni-Netto, R.C. A novel missense mutation p.L76P in the GJB2 gene causing nonsyndromic recessive deafness in a Brazilian family. Braz. J. Med. Biol. Res. 42, 168–171 (2009).
Morris, G.M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Allen, B. Jr. Is CMS's requirement for age-specific clinical evidence before considering coverage for CT colonography reasonable and necessary? J. Am. Coll. Radiol. 6, 606–608 (2009).
Guo, M., Ignatov, M., Musier-Forsyth, K., Schimmel, P. & Yang, X.L. Crystal structure of tetrameric form of human lysyl-tRNA synthetase: implications for multisynthetase complex formation. Proc. Natl. Acad. Sci. USA 105, 2331–2336 (2008).
Ofir-Birin, Y. et al. Structural switch of lysyl-tRNA synthetase between translation and transcription. Mol. Cell 49, 30–42 (2013).
Park, S.G. et al. Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc. Natl. Acad. Sci. USA 102, 6356–6361 (2005).
Kim, J.Y. et al. p38 is essential for the assembly and stability of macromolecular tRNA synthetase complex: implications for its physiological significance. Proc. Natl. Acad. Sci. USA 99, 7912–7916 (2002).
Kwon, N.H. et al. Dual role of methionyl-tRNA synthetase in the regulation of translation and tumor suppressor activity of aminoacyl-tRNA synthetase-interacting multifunctional protein-3. Proc. Natl. Acad. Sci. USA 108, 19635–19640 (2011).
Nam, J.S. et al. Transforming growth factor β subverts the immune system into directly promoting tumor growth through interleukin-17. Cancer Res. 68, 3915–3923 (2008).
Sevenich, L. et al. Transgenic expression of human cathepsin B promotes progression and metastasis of polyoma-middle-T–induced breast cancer in mice. Oncogene 30, 54–64 (2011).
Arguello, F. et al. Incidence and distribution of experimental metastases in mutant mice with defective organ microenvironments (genotypes Sl/Sld and W/Wv). Cancer Res. 52, 2304–2309 (1992).
Pérot, S., Sperandio, O., Miteva, M.A., Camproux, A.C. & Villoutreix, B.O. Druggable pockets and binding site centric chemical space: a paradigm shift in drug discovery. Drug Discov. Today 15, 656–667 (2010).
Cheng, A.C. et al. Structure-based maximal affinity model predicts small-molecule druggability. Nat. Biotechnol. 25, 71–75 (2007).
Fang, P. et al. Structural context for mobilization of a human tRNA synthetase from its cytoplasmic complex. Proc. Natl. Acad. Sci. USA 108, 8239–8244 (2011).
Omar, A., Reusch, U., Knackmuss, S., Little, M. & Weiss, S.F. Anti-LRP/LR-specific antibody IgG1-iS18 significantly reduces adhesion and invasion of metastatic lung, cervix, colon and prostate cancer cells. J. Mol. Biol. 419, 102–109 (2012).
Zuber, C. et al. Invasion of tumorigenic HT1080 cells is impeded by blocking or downregulating the 37-kDa/67-kDa laminin receptor. J. Mol. Biol. 378, 530–539 (2008).
Lukk, M. et al. A global map of human gene expression. Nat. Biotechnol. 28, 322–324 (2010).
Hippo, Y. et al. Global gene expression analysis of gastric cancer by oligonucleotide microarrays. Cancer Res. 62, 233–240 (2002).
Hura, G.L. et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 (2009).
Young, I.S. & Nicholls, D.P. Lipid metabolism. Curr. Opin. Lipidol. 21, 550–551 (2010).
Pettersen, E.F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Soares, M.R., de Paula, F.O., Chaves, M., Assis, N.M. & Chaves Filho, H.D. Patient with Down syndrome and implant therapy: a case report. Braz. Dent. J. 21, 550–554 (2010).
Schmidlin, K. et al. Complication and failure rates in patients treated for chronic periodontitis and restored with single crowns on teeth and/or implants. Clin. Oral Implants Res. 21, 550–557 (2010).
Otani, A. et al. A fragment of human TrpRS as a potent antagonist of ocular angiogenesis. Proc. Natl. Acad. Sci. USA 99, 178–183 (2002).
Cardin, V., Friston, K.J. & Zeki, S. Top-down modulations in the visual form pathway revealed with dynamic causal modeling. Cereb. Cortex 21, 550–562 (2011).
Ford, C.L., Randal-Whitis, L. & Ellis, S.R. Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res. 59, 704–710 (1999).
Kawamata, H., Magrane, J., Kunst, C., King, M.P. & Manfredi, G. Lysyl-tRNA synthetase is a target for mutant SOD1 toxicity in mitochondria. J. Biol. Chem. 283, 28321–28328 (2008).
Kim, E.S. et al. Sphingosine 1-phosphate regulates matrix metalloproteinase-9 expression and breast cell invasion through S1P3-Gαq coupling. J. Cell Sci. 124, 2220–2230 (2011).
Nyberg, P. et al. Endostatin inhibits human tongue carcinoma cell invasion and intravasation and blocks the activation of matrix metalloprotease-2, -9, and -13. J. Biol. Chem. 278, 22404–22411 (2003).
Shenkarev, Z.O. et al. Lipid-protein nanodiscs: possible application in high-resolution NMR investigations of membrane proteins and membrane-active peptides. Biochemistry (Mosc.) 74, 756–765 (2009).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Vranken, W.F. et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins 59, 687–696 (2005).
Kim, K.A. et al. Structure of human PRL-3, the phosphatase associated with cancer metastasis. FEBS Lett. 565, 181–187 (2004).
Putnam, C.D., Hammel, M., Hura, G.L. & Tainer, J.A. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 40, 191–285 (2007).
Konarev, P.V., Volkov, V.V., Sokolova, A.V., Koch, M.H.J. & Svergun, D.I. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 (2003).
Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 25, 495–503 (1992).
Franke, D. & Svergun, D.I. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 42, 342–346 (2009).
Zhang, H.M. et al. Enhanced digestion efficiency, peptide ionization efficiency, and sequence resolution for protein hydrogen/deuterium exchange monitored by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 80, 9034–9041 (2008).
Zhang, H.M., Bou-Assaf, G.M., Emmett, M.R. & Marshall, A.G. Fast reversed-phase liquid chromatography to reduce back exchange and increase throughput in H/D exchange monitored by FT-ICR mass spectrometry. J. Am. Soc. Mass Spectrom. 20, 520–524 (2009).
Zhang, H.M. et al. Simultaneous reduction and digestion of proteins with disulfide bonds for hydrogen/deuterium exchange monitored by mass spectrometry. Anal. Chem. 82, 1450–1454 (2010).
Schaub, T.M. et al. High-performance mass spectrometry: Fourier transform ion cyclotron resonance at 14.5 Tesla. Anal. Chem. 80, 3985–3990 (2008).
Kazazic, S. et al. Automated data reduction for hydrogen/deuterium exchange experiments, enabled by high-resolution Fourier transform ion cyclotron resonance mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 550–558 (2010).
Zhang, Z., Li, W., Logan, T.M., Li, M. & Marshall, A.G. Human recombinant [C22A] FK506-binding protein amide hydrogen exchange rates from mass spectrometry match and extend those from NMR. Protein Sci. 6, 2203–2217 (1997).
Beebe, K., Waas, W., Druzina, Z., Guo, M. & Schimmel, P. A universal plate format for increased throughput of assays that monitor multiple aminoacyl transfer RNA synthetase activities. Anal. Biochem. 368, 111–121 (2007).
Acknowledgements
This work was supported by the Global Frontier Project (grants NRF-M1AXA002-2010-0029785 (S.K.), NRF-M1AXA002-2010-0029765 (Y.H.J.), NRF-2012M3A6A4054271 (J.W.L.) and NRF- 2012-054237 (B.W.H.)), the World Class University (grant R31-2008-000-10086-0 (G.H.)), the Proteogenomic Research Program (2012M3A9B9036679 (C.L.)) and by the National Research Foundation funded by the Ministry of Education, Science and Technology of Korea (grant ROA-2012-0006262 (A.M.)). This study was also funded by the Ministry for Health and Welfare Affairs of Korea through the Korea Healthcare Technology Research and Development Project (A092255 (D.-H.N.)) and by a grant from Gyeonggi Research Development Program (S.K.). This work was supported in part by grants from the US National Institutes of Health (GM100136 (M.G.)); the US National Science Foundation Division of Materials Research through DMR-11-57490 (A.G. Marshall, Florida State University); by the state of Florida (M.G.) and by a Kimmel Scholar Award for Cancer Research (M.G.). We appreciate A.G. Marshall for supporting the HDX-MS program and facility, M.-S. Seok (Korea University) for the preparation of the nanodisc and S.W. Lee (Sungkyunkwan University) for X-ray analysis.
Author information
Authors and Affiliations
Contributions
D.G.K., J.Y.L. and S.K. designed experiments. D.G.K., J.Y.L., P.F., Q.Z., J.W., H.Y.C., A.U.M., J.W.C., H.W.K., J.E.J., W.K., H.Y., M.-S.L., E.-S.K., E.J.K., J.S.Y., W.S.Y. and J.S.Y. performed experiments. D.G.K., J.Y.L., N.H.K., N.L.Y., M.G., Y.H.J., H.W.K., Y.H., D.-H.N., J.W.L., A.M., J.M.H., G.H., C.L. and S.K. analyzed the data. Y.H., K.K., Y.M., D.K., K.H.R. and B.W.H. provided materials and supported experiments. D.G.K., N.H.K., M.G., Y.H.J., Y.H. and S.K. wrote the manuscript. D.G.K., J.Y.L., N.H.K., M.G., Y.H.J., H.W.K., Y.H., D.-H.N. and S.K. reviewed the manuscript. D.G.K., J.Y.L., N.H.K., M.G., Y.H.J., G.H., M.W.W. and S.K. discussed the results and commented.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Results, Supplementary Tables 1–4, Supplementary Figures 1–24 and Supplementary Note. (PDF 14418 kb)
Rights and permissions
About this article
Cite this article
Kim, D., Lee, J., Kwon, N. et al. Chemical inhibition of prometastatic lysyl-tRNA synthetase–laminin receptor interaction. Nat Chem Biol 10, 29–34 (2014). https://doi.org/10.1038/nchembio.1381
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1381
This article is cited by
-
Anti-apoptotic Splicing Variant of AIMP2 Recover Mutant SOD1-Induced Neuronal Cell Death
Molecular Neurobiology (2023)
-
Functional and pathologic association of aminoacyl-tRNA synthetases with cancer
Experimental & Molecular Medicine (2022)
-
Human lysyl-tRNA synthetase evolves a dynamic structure that can be stabilized by forming complex
Cellular and Molecular Life Sciences (2022)
-
Aminoacyl-tRNA synthetases as therapeutic targets
Nature Reviews Drug Discovery (2019)
-
Biocon's target factory
Nature Biotechnology (2018)