Abstract
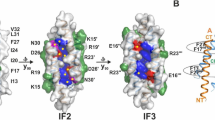
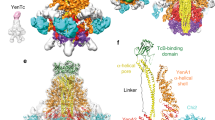
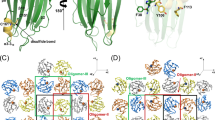
Human pathogens often produce soluble protein toxins that generate pores inside membranes, resulting in the death of target cells and tissue damage. In pathogenic amoebae, this has been exemplified with amoebapores of the enteric protozoan parasite Entamoeba histolytica. Here we characterize acanthaporin, to our knowledge the first pore-forming toxin to be described from acanthamoebae, which are free-living, bacteria-feeding, unicellular organisms that are opportunistic pathogens of increasing importance and cause severe and often fatal diseases. We isolated acanthaporin from extracts of virulent Acanthamoeba culbertsoni by tracking its pore-forming activity, molecularly cloned the gene of its precursor and recombinantly expressed the mature protein in bacteria. Acanthaporin was cytotoxic for human neuronal cells and exerted antimicrobial activity against a variety of bacterial strains by permeabilizing their membranes. The tertiary structures of acanthaporin's active monomeric form and inactive dimeric form, both solved by NMR spectroscopy, revealed a currently unknown protein fold and a pH-dependent trigger mechanism of activation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Leippe, M. Antimicrobial and cytolytic polypeptides of amoeboid protozoa—effector molecules of primitive phagocytes. Dev. Comp. Immunol. 23, 267–279 (1999).
Herbst, R. et al. Pore-forming polypeptides of the pathogenic protozoon Naegleria fowleri. J. Biol. Chem. 277, 22353–22360 (2002).
Herbst, R., Marciano-Cabral, F. & Leippe, M. Antimicrobial and pore-forming peptides of free-living and potentially highly pathogenic Naegleria fowleri are released from the same precursor molecule. J. Biol. Chem. 279, 25955–25958 (2004).
Hecht, O. et al. Solution structure of the pore-forming protein of Entamoeba histolytica. J. Biol. Chem. 279, 17834–17841 (2004).
Kolter, T., Winau, F., Schaible, U.E., Leippe, M. & Sandhoff, K. Lipid-binding proteins in membrane digestion, antigen presentation, and antimicrobial defense. J. Biol. Chem. 280, 41125–41128 (2005).
Liepinsh, E., Andersson, M., Ruysschaert, J.M. & Otting, G. Saposin fold revealed by the NMR structure of NK-lysin. Nat. Struct. Biol. 4, 793–795 (1997).
Anderson, D.H. et al. Granulysin crystal structure and a structure-derived lytic mechanism. J. Mol. Biol. 325, 355–365 (2003).
Leippe, M., Bruhn, H., Hecht, O. & Grötzinger, J. Ancient weapons: the three-dimensional structure of amoebapore A. Trends Parasitol. 21, 5–7 (2005).
Marciano-Cabral, F. & Cabral, G. Acanthamoeba spp. as agents of disease in humans. Clin. Microbiol. Rev. 16, 273–307 (2003).
Martinez, A.J. Free-living amebas: infection of the central nervous system. Mt. Sinai J. Med. 60, 271–278 (1993).
Visvesvara, G.S. Amebic meningoencephalitides and keratitis: challenges in diagnosis and treatment. Curr. Opin. Infect. Dis. 23, 590–594 (2010).
Greub, G. & Raoult, D. Microorganisms resistant to free-living amoebae. Clin. Microbiol. Rev. 17, 413–433 (2004).
Ellman, G.L. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 (1959).
Holm, L., Kaariainen, S., Rosenstrom, P. & Schenkel, A. Searching protein structure databases with DaliLite v.3. Bioinformatics 24, 2780–2781 (2008).
Madej, T., Gibrat, J.F. & Bryant, S.H. Threading a database of protein cores. Proteins 23, 356–369 (1995).
Gibrat, J.F., Madej, T. & Bryant, S.H. Surprising similarities in structure comparison. Curr. Opin. Struct. Biol. 6, 377–385 (1996).
Andrä, J., Herbst, R. & Leippe, M. Amoebapores, archaic effector peptides of protozoan origin, are discharged into phagosomes and kill bacteria by permeabilizing their membranes. Dev. Comp. Immunol. 27, 291–304 (2003).
Yeaman, M.R. & Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 55, 27–55 (2003).
Rötzschke, O., Lau, J.M., Hofstatter, M., Falk, K. & Strominger, J.L. A pH-sensitive histidine residue as control element for ligand release from HLA-DR molecules. Proc. Natl. Acad. Sci. USA 99, 16946–16950 (2002).
Baumann, G. & Mueller, P. A molecular model of membrane excitability. J. Supramol. Struct. 2, 538–557 (1974).
Shai, Y. Mechanism of the binding, insertion and destabilization of phospholipid bilayer membranes by α-helical antimicrobial and cell non-selective membrane-lytic peptides. Biochim. Biophys. Acta 1462, 55–70 (1999).
Yang, L., Harroun, T.A., Weiss, T.M., Ding, L. & Huang, H.W. Barrel-stave model or toroidal model? A case study on melittin pores. Biophys. J. 81, 1475–1485 (2001).
Anderluh, G. & Lakey, J.H. Disparate proteins use similar architectures to damage membranes. Trends Biochem. Sci. 33, 482–490 (2008).
Jenssen, H., Hamill, P. & Hancock, R.E. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19, 491–511 (2006).
Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 415, 389–395 (2002).
Gutsmann, T. et al. Interaction of amoebapores and NK-lysin with symmetric phospholipid and asymmetric lipopolysaccharide/phospholipid bilayers. Biochemistry 42, 9804–9812 (2003).
Leippe, M., Andrä, J., Nickel, R., Tannich, E. & Müller-Eberhard, H.J. Amoebapores, a family of membranolytic peptides from cytoplasmic granules of Entamoeba histolytica: isolation, primary structure, and pore formation in bacterial cytoplasmic membranes. Mol. Microbiol. 14, 895–904 (1994).
Schägger, H. & von Jagow, G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368 (1987).
Frohman, M.A., Dush, M.K. & Martin, G.R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. USA 85, 8998–9002 (1988).
Scotto-Lavino, E., Du, G. & Frohman, M.A. 3′ end cDNA amplification using classic RACE. Nat. Protoc. 1, 2742–2745 (2006).
LaVallie, E.R. et al. A thioredoxin gene fusion expression system that circumvents inclusion body formation in the E. coli cytoplasm. Bio/Technology 11, 187–193 (1993).
Dingley, A.J., Lorenzen, I. & Grötzinger, J. NMR analysis of viral protein structures. Methods Mol. Biol. 451, 441–462 (2008).
Markley, J.L. et al. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J. Mol. Biol. 280, 933–952 (1998).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Goddard, T. & Kneller, D. SPARKY 3 (University of San Francisco, California, USA.)
Johnson, B.A. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 278, 313–352 (2004).
Güntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 (2004).
Schwieters, C.D., Kuszewski, J.J., Tjandra, N. & Clore, G.M. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 160, 65–73 (2003).
Linge, J.P., Williams, M.A., Spronk, C.A., Bonvin, A.M. & Nilges, M. Refinement of protein structures in explicit solvent. Proteins 50, 496–506 (2003).
Koradi, R., Billeter, M. & Wuthrich, K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 (1996).
Andrä, J. & Leippe, M. Pore-forming peptide of Entamoeba histolytica. Significance of positively charged amino acid residues for its mode of action. FEBS Lett. 354, 97–102 (1994).
Montal, M. & Mueller, P. Formation of bimolecular membranes from lipid monolayers and a study of their electrical properties. Proc. Natl. Acad. Sci. USA 69, 3561–3566 (1972).
Dougherty, R.M., Galli, C., Ferro-Luzzi, A. & Iacono, J.M. Lipid and phospholipid fatty acid composition of plasma, red blood cells, and platelets and how they are affected by dietary lipids: a study of normal subjects from Italy, Finland, and the USA. Am. J. Clin. Nutr. 45, 443–455 (1987).
Bruhn, H., Riekens, B., Berninghausen, O. & Leippe, M. Amoebapores and NK-lysin, members of a class of structurally distinct antimicrobial and cytolytic peptides from protozoa and mammals: a comparative functional analysis. Biochem. J. 375, 737–744 (2003).
Boman, H.G., Nilsson-Faye, I., Paul, K. & Rasmuson, T. Jr. Insect immunity. I. Characteristics of an inducible cell-free antibacterial reaction in hemolymph of Samia cynthia pupae. Infect. Immun. 10, 136–145 (1974).
Leippe, M., Ebel, S., Schoenberger, O.L., Horstmann, R.D. & Müller-Eberhard, H.J. Pore-forming peptide of pathogenic Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 88, 7659–7663 (1991).
Acknowledgements
The authors acknowledge support in measuring the 750-MHz NMR spectra at The Netherlands Foundation for Chemical Research (SON) NMR Large Scale Facility in Utrecht, The Netherlands, funded by the European Union (contract number RII3-026145). We thank F. Buck, Institute for Cell Biochemistry and Clinical Neurobiology, University of Hamburg, for protein sequencing, C. Ott for expert technical assistance during purification of the natural acanthaporin and H. Ließegang for technical help during antibacterial activity testing. This work was supported by German Research Council (DFG) grant LE 1075/2-4 to M.L. and J.G. and the excellence cluster 306 'Inflammation at Interfaces'.
Author information
Authors and Affiliations
Contributions
M.L. conceived the study and purified the natural protein. M.M. performed the majority of the experiments. F.D.S., R.W., A.J.D. and H.W. contributed to structure determination and data analysis. C.-W.H. and A.T. assigned the disulfide bond connectivity. A.K. and T.G. completed the planar lipid bilayer experiments. M.S., R.H. and I.L. performed molecular biology and initial protein expression. M.S. contributed to antibacterial assays. F.M.-C. cultured and passaged the amoebae and made the amoebic extracts. C.G. performed the initial mass spectrometry measurements of the protein. M.L. and J.G. directed experiments. M.M. and M.L. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Note and Supplementary Results (PDF 1648 kb)
Rights and permissions
About this article
Cite this article
Michalek, M., Sönnichsen, F., Wechselberger, R. et al. Structure and function of a unique pore-forming protein from a pathogenic acanthamoeba. Nat Chem Biol 9, 37–42 (2013). https://doi.org/10.1038/nchembio.1116
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.1116
This article is cited by
-
Proteases of Acanthamoeba
Parasitology Research (2024)
-
War of the microbial world: Acanthamoeba spp. interactions with microorganisms
Folia Microbiologica (2021)
-
Novel hemagglutinating, hemolytic and cytotoxic activities of the intermediate subunit of Entamoeba histolytica lectin
Scientific Reports (2015)
-
Superdiffusion dominates intracellular particle motion in the supercrowded cytoplasm of pathogenic Acanthamoeba castellanii
Scientific Reports (2015)
-
Cytotoxic activity and degradation patterns of structural proteins by corneal isolates of Acanthamoeba spp
Graefe's Archive for Clinical and Experimental Ophthalmology (2015)