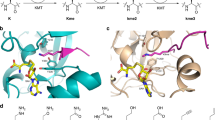
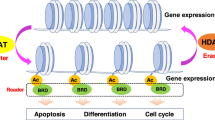
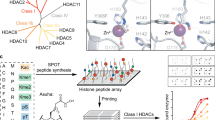
Abstract
The histone-modifying enzymes that catalyze reversible lysine acetylation and methylation are central to the epigenetic regulation of chromatin remodeling. From the early discovery of histone deacetylase inhibitors to the more recent identification of histone demethylase blockers, chemical approaches offer increasingly sophisticated tools for the investigation of the structure and function of these lysine-modifying enzymes. This review summarizes progress to date on compounds identified from screens or by design that can modulate the activity of classical histone deacetylases, sirtuins, histone acetyltransferases, histone methyltransferases and histone demethylases. We highlight applications of compounds to mechanistic and functional studies involving these enzymes and discuss future challenges regarding target specificity and general utility.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Goldberg, A.D., Allis, C.D. & Bernstein, E. Epigenetics: a landscape takes shape. Cell 128, 635–638 (2007).
Taverna, S.D., Li, H., Ruthenburg, A.J., Allis, C.D. & Patel, D.J. How chromatin-binding modules interpret histone modifications. Nat. Struct. Mol. Biol. 14, 1025–1040 (2007).
Allfrey, V.G., Faulkner, R. & Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51, 786–794 (1964).
Murray, K. The occurrence of ε-N-methyl lysine in histones. Biochemistry 3, 10–15 (1964).
Gershey, E.L., Vidali, G. & Allfrey, V.G. Chemical studies of histone acetylation. The occurrence of ε-N-acetyllysine in the f2a1 histone. J. Biol. Chem. 243, 5018–5022 (1968).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Brownell, J.E. et al. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84, 843–851 (1996).
Taunton, J., Hassig, C.A. & Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 (1996).
Vetting, M.W. et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 433, 212–226 (2005).
Yoshida, M., Kijima, M., Akita, M. & Beppu, T. Potent and specific inhibition of mammalian histone deacetylase in vivo and in vitro by trichostatin A. J. Biol. Chem. 265, 17174–17179 (1990).
Kijima, M., Yoshida, M., Sugita, K., Horinouchi, S. & Beppu, T. Trapoxin, an antitumor cyclic tetrapeptide, is an inhibitor of mammalian histone deacetylase. J. Biol. Chem. 268, 22429–22435 (1993).
Finnin, M.S. et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401, 188–193 (1999).
Itoh, Y., Suzuki, T. & Miyata, N. Isoform-selective histone deacetylase inhibitors. Curr. Pharm. Des. 14, 529–544 (2008).
Imai, S., Armstrong, C.M., Kaeberlein, M. & Guarente, L. Transcriptional silencing and longetivity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403, 795–800 (2000).
Tanny, J.C., Dowd, G.J., Huang, J., Hilz, H. & Moazed, D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell 99, 735–745 (1999).
Blander, G. & Guarente, L. The Sir2 family of protein deacetylases. Annu. Rev. Biochem. 73, 417–435 (2004).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406, 593–599 (2000).
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6, 838–849 (2005).
Shi, Y. et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 (2004).
Tsukada, Y. et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 (2006).
Culhane, J.C. & Cole, P.A. LSD1 and the chemistry of histone demethylation. Curr. Opin. Chem. Biol. 11, 561–568 (2007).
Paris, M., Porcelloni, M., Binaschi, M. & Fattori, D. Histone deacetylase inhibitors: from bench to clinic. J. Med. Chem. 51, 1505–1529 (2008).
Vannini, A. et al. Crystal structure of a eukaryotic zinc-dependent histone deacetylase, human HDAC8, complexed with a hydroxamic acid inhibitor. Proc. Natl. Acad. Sci. USA 101, 15064–15069 (2004).
Schuetz, A. et al. Human HDAC7 harbors a class IIa histone deacetylase-specific zinc binding motif and cryptic deacetylase activity. J. Biol. Chem. 283, 11355–11363 (2008).
Jones, P.A. & Baylin, S.B. The epigenomics of cancer. Cell 128, 683–692 (2007).
Wong, J.C., Hong, R. & Schreiber, S.L. Structural basis for in-cell histone deacetylase paralog selectivity. J. Am. Chem. Soc. 125, 5586–5587 (2003).
Estiu, G. et al. Structural origin of selectivity in class II-selective histone deacetylase inhibitors. J. Med. Chem. 51, 2898–2906 (2008).
Hideshima, T. et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proc. Natl. Acad. Sci. USA 102, 8567–8572 (2005).
Tran, A.D. et al. DAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. J. Cell Sci. 120, 1469–1479 (2007).
Salisbury, C.M. & Cravatt, B.F. Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes. J. Am. Chem. Soc. 130, 2184–2194 (2008).
Guarente, L. & Picard, F. Calorie restriction–the SIR2 connection. Cell 120, 473–482 (2005).
Guarente, L. Sirtuins in aging and disease. Cold Spring Harb. Symp. Quant. Biol. 72, 483–488 (2007).
Wade, N. New hints seen that red wine may slow aging. New York Times 4, June 2008A1.
Howitz, K.T. et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 (2003).
Borra, M.T., Smith, B.C. & Denu, J.M. Mechanism of human SirT1 activation by resveratrol. J. Biol. Chem. 280, 17187–17195 (2005).
Kaeberlein, M. et al. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 280, 17038–17045 (2005).
Lagouge, M. et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell 127, 1109–1122 (2006).
Szewczuk, L.M., Lee, S.H., Blair, I.A. & Penning, T.M. Vinferin formation by COX-1: evidence for radical intermediates during co-oxidation of resveratrol. J. Nat. Prod. 68, 36–42 (2005).
Milne, J.C. et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450, 712–716 (2007).
Hoff, K.G., Avalos, J.L., Sens, K. & Wolberger, C. Insights into the sirtuin mechanism from ternary complexes containing NAD+ and acetylated peptide. Structure 14, 1231–1240 (2006).
Liu, Y. et al. A fasting inducible acetylase/deacetylase switch modulates gluconeogenesis through activator-coactivator exchange. Nature (in the press).
Schmidt, M.T., Smith, B.C., Jackson, M.D. & Denu, J.M. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 279, 40122–40129 (2004).
Luo, J. et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 (2001).
Fatkins, D.G., Monnot, A.D. & Zheng, W. Nε-thioacetyl-lysine: a multi-facet functional probe for enzymatic protein lysine Nε-deacetylation. Bioorg. Med. Chem. Lett. 16, 3651–3656 (2006).
Smith, B.C. & Denu, J.M. Acetyl-lysine analog peptides as mechanistic probes of protein deacetylases. J. Biol. Chem. 282, 37256–37265 (2007).
Bedalov, A., Gatbonton, T., Irvine, W.P., Gottschling, D.E. & Simon, J.A. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. USA 98, 15113–15118 (2001).
Grozinger, C.M., Chao, E.D., Blackwell, H.E., Moazed, D. & Schreiber, S.L. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J. Biol. Chem. 276, 38837–38843 (2001).
Hirao, M. et al. Identification of selective inhibitors of NAD-dependent deacetylases using phenotypic screens in yeast. J. Biol. Chem. 278, 52773–52782 (2003).
Zhao, Y., Dai, X., Blackwell, H.E., Schreiber, S.L. & Chory, J. Sir1, an upstream component in auxin signaling identified by chemical genetics. Science 301, 1107–1110 (2003).
Araki, T., Sasaki, Y. & Milbrandt, J. Increased nuclear NAD biosynthesis and SirT1 activation prevent axonal degeneration. Science 305, 1010–1013 (2004).
Alcendor, R.R., Kirshenbaum, L.A., Imai, S., Vatner, S.F. & Sadoshima, J. Silent information regulator 2a, a longevity factor and class III histone deacetylase, is an essential endogenous apoptosis inhibitor in cardiac myocytes. Circ. Res. 95, 971–980 (2004).
Outeiro, T.F. et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson's disease. Science 317, 516–519 (2007).
Swaminathan, V. et al. Small molecule modulators in epigenetics: implications in gene expression and therapeutics. Subcell. Biochem. 41, 397–428 (2007).
Zheng, Y. et al. Selective HAT inhibitors as mechanistic tools for protein acetylation. Methods Enzymol. 376, 188–199 (2004).
Stimson, L. et al. Isothiazolones as inhibitors of PCAF and p300 histone acetyltransferase activity. Mol. Cancer Ther. 4, 1521–1532 (2005).
Mantelingu, K. et al. Specific inhibition of p300-HAT alters global gene expression and represses HIV replication. Chem. Biol. 14, 645–657 (2007).
Morimoto, T. et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J. Clin. Invest. 118, 868–878 (2008).
Chase, J.F.A. & Tubbs, P.K. Conditions for the self-catalysed inactivation of carnitine acetyltransferase. Biochem. J. 111, 225–235 (1969).
Parang, K. et al. Mechanism-based design of a protein kinase inhibitor. Nat. Struct. Biol. 8, 37–41 (2001).
Lau, O.D. et al. HATs off: selective synthetic inhibitors of the histone acetyltransferases p300 and PCAF. Mol. Cell 5, 589–595 (2000).
Lau, O.D. et al. PCAF Histone acetyltransferase processing of a peptide substrate: kinetic analysis of the catalytic mechanism. J. Biol. Chem. 275, 21953–21959 (2000).
Poux, A.N., Cebrat, M., Kim, C.M., Cole, P.A. & Marmorstein, R. Structure of the GCN5 histone acetyltransferase bound to a bisubstrate inhibitor. Proc. Natl. Acad. Sci. USA 99, 14065–14070 (2002).
Zheng, Y. et al. Fluorescence analysis of a dynamic loop in the PCAF/GCN5 histone acetyltransferase. Biochemistry 44, 10501–10509 (2005).
Thompson, P.R., Kurooka, H., Nakatani, Y. & Cole, P.A. Transcriptional coactivator protein p300. Kinetic characterization of its histone acetyltransferase activity. J. Biol. Chem. 276, 33721–33729 (2001).
Liu, X. et al. The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451, 846–850 (2008).
Zheng, Y. et al. Synthesis and evaluation of a potent and selective cell-permeable p300 histone acetyltransferase inhibitor. J. Am. Chem. Soc. 127, 17182–17183 (2005).
Guidez, F. et al. Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic zinc finger protein. Mol. Cell. Biol. 25, 5552–5566 (2005).
Cleary, J. et al. Acetylation by P/CAF drives DEK into interchromatin granule clusters. J. Biol. Chem. 280, 31760–31767 (2005).
Kenneth, N.S. et al. Activation of Pol III transcription by c-Myc involves selective acetylation of histone H3 and recruitment of TRAPP, GCN5, and TFIIIB. Proc. Natl. Acad. Sci. USA 104, 14917–14922 (2007).
Huang, S. Histone methyltransferases, diet nutrients and tumour suppressors. Nat. Rev. Cancer 2, 469–476 (2002).
Dirk, L.M. et al. Kinetic manifestation of processivity during multiple methylations catalyzed by SET domain protein methyltransferases. Biochemistry 46, 3905–3915 (2007).
Diehl, F., Rössig, L., Zeiher, A.M., Dimmeler, S. & Urbich, C. The histone methyltransferase MLL is an upstream regulator of endothelial-cell sprout formation. Blood 109, 1472–1478 (2007).
Greiner, D., Bonaldi, T., Eskeland, R., Roemer, E. & Imhof, A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat. Chem. Biol. 1, 143–145 (2005).
Kubicek, S. et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481 (2007).
Isham, C.R. et al. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood 109, 2579–2588 (2007).
Walsh, C.T. Suicide substrates, mechanism-based enzyme inactivators: recent developments. Annu. Rev. Biochem. 53, 493–535 (1984).
Edmondson, D.E., Mattevi, A., Binda, C., Li, M. & Hubálek, F. Structure and mechanism of monoamine oxidase. Curr. Med. Chem. 11, 1983–1993 (2004).
Metzger, E. et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 (2005).
Forneris, F., Binda, C., Vanoni, M.A., Battaglioli, E. & Mattevi, A. Human histone demethylase LSD1 reads the histone code. J. Biol. Chem. 280, 41360–41365 (2005).
Lee, M.G., Wynder, C., Schmidt, D.M., McCafferty, D.G. & Shiekhatter, R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem. Biol. 13, 563–567 (2006).
Schmidt, D.M. & McCafferty, D.G. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry 46, 4408–4416 (2007).
Yang, M. et al. Structural basis for inhibition of the LSD1 histone demethylase by the antidepressant trans-2-Phenylcyclopropylamine. Biochemistry 46, 8058–8065 (2007).
Mimasu, S., Sengoku, T., Fukuzawa, S., Umehara, T. & Yokoyama, S. Crystal structure of histone demethylase LSD1 and tranylcypromine at 2.25 A. Biochem. Biophys. Res. Commun. 366, 15–22 (2008).
Culhane, J.C. et al. A mechanism-based inactivator for histone demethylase LSD1. J. Am. Chem. Soc. 128, 4536–4537 (2006).
Szewczuk, L.M. et al. Mechanistic analysis of a suicide inactivator of histone demethylase LSD1. Biochemistry 46, 6892–6902 (2007).
Yang, M. et al. Structural basis for histone demethylation by LSD1 revealed by suicide inactivation. Nat. Struct. Mol. Biol. 14, 535–539 (2007).
Ramanathan, S.K. et al. Modular synthesis of cyclic peptidomimetics inspired by γ-turns. Org. Lett. 7, 1059–1062 (2005).
Forneris, F., Binda, C., Adamo, A., Battaglioli, E. & Mattevi, A. Structural basis of LSD1-CoREST selectivity in histone H3 recognition. J. Biol. Chem. 282, 20070–20074 (2007).
Huang, Y. et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc. Natl. Acad. Sci. USA 104, 8023–8028 (2007).
Osborne, T., Roska, R.L., Rajski, S.R. & Thompson, P.R. In situ generation of a bisubstrate analogue for protein arginine methyltransferase 1. J. Am. Chem. Soc. 130, 4574–4575 (2008).
Luo, Y., Knuckley, B., Bhatia, M., Pellechia, P.J. & Thompson, P.R. Activity-based protein profiling reagents for protein arginine deiminase 4 (PAD4): synthesis and in vitro evaluation of a fluorescently labeled probe. J. Am. Chem. Soc. 128, 14468–14469 (2006).
Yu, M., de Carvalho, L.P., Sun, G. & Blanchard, J.S. Activity-based substrate profiling for Gcn5-related N-acetyltransferases: the use of chloroacetyl-coenzyme A to identify protein substrates. J. Am. Chem. Soc. 128, 15356–15357 (2006).
Hwang, Y. et al. Selective chemical probe for coenzyme A-requiring enzymes. Angew. Chem. Int. Edn Engl. 46, 7621–7624 (2007).
Sachchidanand et al. Target structure-based discovery of small molecules that block human p53 and CREB binding protein association. Chem. Biol. 13, 81–90 (2006).
Shogren-Knaak, M. et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 (2006).
Thompson, P.R. et al. Regulation of p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 11, 308–315 (2004).
Simon, M.D. et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128, 1003–1012 (2007).
McGinty, R.K., Kim, J., Chatterjee, C., Roeder, R.G. & Muir, T.W. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature 453, 812–816 (2008).
Lin, C.W., Jao, C.Y. & Ting, A.Y. Genetically encoded fluorescent reporters of histone methylation in living cell. J. Am. Chem. Soc. 126, 5982–5983 (2004).
Shogren-Knaak, M.A., Alaimo, P.J. & Shokat, K.M. Recent advances in chemical approaches to the study of biological systems. Annu. Rev. Cell Biol. 17, 405–433 (2001).
Qiao, Y., Molina, H., Pandey, A., Zhang, J. & Cole, P.A. Chemical rescue of a mutant enzyme in living cells. Science 311, 1293–1297 (2006).
Acknowledgements
I am grateful to members of my group past and present, as well as many collaborators in this field for many helpful discussions and for their key roles in the work cited. I also thank the US National Institutes of Health, the Flight Attendant Medical Research Institute and the Kaufman Foundation for support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cole, P. Chemical probes for histone-modifying enzymes. Nat Chem Biol 4, 590–597 (2008). https://doi.org/10.1038/nchembio.111
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.111
This article is cited by
-
CRNDE acts as an epigenetic modulator of the p300/YY1 complex to promote HCC progression and therapeutic resistance
Clinical Epigenetics (2022)
-
Identification of Four Enhancer-Associated Genes as Risk Signature for Diffuse Glioma Patients
Journal of Molecular Neuroscience (2022)
-
The histone methyltransferase inhibitor A-366 enhances hemoglobin expression in erythroleukemia cells upon co‐exposure with chemical inducers in culture
Journal of Biological Research-Thessaloniki (2021)
-
Materials control of the epigenetics underlying cell plasticity
Nature Reviews Materials (2020)
-
Structure-guided development of YEATS domain inhibitors by targeting π-π-π stacking
Nature Chemical Biology (2018)