Abstract
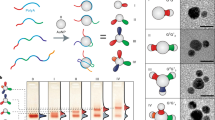
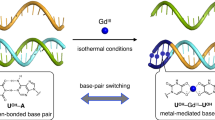
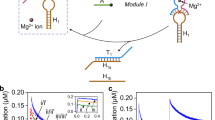
The universal genetic code relies on two hydrogen-bonded Watson–Crick base pairs that can form 64 triplet codons. This places a limit on the number of amino acids that can be encoded, which has motivated efforts to create synthetic base pairs that are orthogonal to the natural ones. An additional base pair would result in another 61 triplet codons. Artificial organic base pairs have been described in enzymatic incorporation studies, and inorganic T–Hg–T and C–Ag–C base pairs have been reported to form in primer extension studies. Here, we demonstrate a metal base pair that is fully orthogonal and can be replicated, and can even be amplified by polymerase chain reaction in the presence of the canonical pairs dA:dT and dG:dC. Crystal structures of a dS–Cu–dS base pair inside a polymerase show that reversible chemistry is possible directly inside the polymerase, which enables the efficient copying of the inorganic crosslink. The results open up the possibility of replicating and amplifying artificial inorganic DNA nanostructures by extending the genetic alphabet.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Giuseppone, N., Schmitt, J.-L. & Lehn, J.-M. Generation of dynamic constitutional diversity and driven evolution in helical molecular strands under Lewis acid catalyzed component exchange. Angew. Chem. Int. Ed. 43, 4902–4906 (2004).
Lehn, J.-M. From supramolecular chemistry towards constitutional dynamic chemistry and adaptive chemistry. Chem. Soc. Rev. 36, 151–160 (2007).
Lu, Y. & Liu, J. Functional DNA nanotechnology: emerging applications of DNAzymes and aptamers. Curr. Opin. Biotechnol. 17, 580–588 (2006).
Niemeyer, C. M. Semisynthetic DNA–protein conjugates for biosensing and nanofabrication. Angew. Chem. Int. Ed. 49, 1200–1216 (2010).
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003).
Switzer, C., Moroney, S. E. & Benner, S. A. Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 111, 8322–8323 (1989).
Crick, F. H. C., Barnett, L., Brenner, S. & Watts-Tobin, R. J. General nature of the genetic code for proteins. Nature 192, 1227–1232 (1961).
Kool, E. T. Synthetically modified DNAs as substrates for polymerases. Curr. Opin. Chem. Biol. 4, 602–608 (2000).
Henry, A. A. & Romesberg, F. E. Beyond A, C, G and T: augmenting Nature's alphabet. Curr. Opin. Chem. Biol. 7, 727–733 (2003).
Hirao, I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 10, 622–627 (2006).
Kool, E. T. Replacing the nucleobases in DNA with designer molecules. Acc. Chem. Res. 35, 936–943 (2002).
Kool, E. T., Morales, J. C. & Guckian, K. M. Mimicking the structure and function of DNA: insights into DNA stability and replication. Angew. Chem. Int. Ed. 39, 990–1009 (2000).
Krueger, A. T. & Kool, E. T. Redesigning the architecture of the base pair: toward biochemical and biological function of new genetic sets. Chem. Biol. 16, 242–248 (2009).
Switzer, C. Y., Moroney, S. E. & Benner, S. A. Enzymic recognition of the base pair between isocytidine and isoguanosine. Biochemistry 32, 10489–10496 (1993).
Killelea, T. et al. Probing the interaction of archaeal DNA polymerases with deaminated bases using X-ray crystallography and non-hydrogen bonding isosteric base analogues. Biochemistry 49, 5772–5781 (2010).
Kimoto, M., Mitsui, T., Yokoyama, S. & Hirao, I. A unique fluorescent base analogue for the expansion of the genetic alphabet. J. Am. Chem. Soc. 132, 4988–4989 (2010).
Leconte, A. M. et al. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J. Am. Chem. Soc. 130, 2336–2343 (2008).
Lu, H., Krueger, A. T., Gao, J., Liu, H. & Kool, E. T. Toward a designed genetic system with biochemical function: polymerase synthesis of single and multiple size-expanded DNA base pairs. Org. Biomol. Chem. 8, 2704–2710 (2010).
Matsuda, S. et al. Efforts toward expansion of the genetic alphabet: structure and replication of unnatural base pairs. J. Am. Chem. Soc. 129, 10466–10473 (2007).
Moran, S., Ren, R. X. F. & Kool, E. T. A thymidine triphosphate shape analog lacking Watson–Crick pairing ability is replicated with high sequence selectivity. Proc. Natl Acad. Sci. USA 94, 10506–10511 (1997).
Moran, S., Ren, R. X. F., Rumney, S. I. V. & Kool, E. T. Difluorotoluene, a nonpolar isostere for thymine, codes specifically and efficiently for adenine in DNA replication. J. Am. Chem. Soc. 119, 2056–2057 (1997).
Hirao, I. et al. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nature Methods 3, 729–735 (2006).
Hirao, I., Mitsui, T., Kimoto, M. & Yokoyama, S. Development of an unnatural base pair for efficient PCR amplification. Nucleic Acids Symp. Ser. 51, 9–10 (2007).
Johnson, S. C., Sherrill, C. B., Marshall, D. J., Moser, M. J. & Prudent, J. R. A third base pair for the polymerase chain reaction: inserting isoC and isoG. Nucleic Acids Res. 32, 1937–1941 (2004).
Yang, Z., Chen, F., Chamberlin, S. G. & Benner, S. A. Expanded genetic alphabets in the polymerase chain reaction. Angew. Chem. Int. Ed. 49, 177–180 (2010).
Piccirilli, J. A., Krauch, T., Moroney, S. E. & Benner, S. A. Enzymic incorporation of a new base pair into DNA and RNA extends the genetic alphabet. Nature 343, 33–37 (1990).
Lutz, M. J., Horlacher, J. & Benner, S. A. Recognition of a non-standard base pair by thermostable DNA polymerases. Bioorg. Med. Chem. Lett. 8, 1149–1152 (1998).
Urata, H., Yamaguchi, E., Funai, T., Matsumura, Y. & Wada, S.-i. Incorporation of thymine nucleotides by DNA polymerases through T-HgII-T base pairing. Angew. Chem. Int. Ed. 49, 6516–6519 (2010).
Katz, S. The reversible reaction of sodium thymonucleate and mercuric chloride. J. Am. Chem. Soc. 74, 2238–2245 (1952).
Kuklenyik, Z. & Marzilli, L. G. Mercury(II) site-selective binding to a DNA hairpin. Relationship of sequence-dependent intra- and interstrand cross-linking to the hairpin–duplex conformational transition. Inorg. Chem. 35, 5654–5662 (1996).
Miyake, Y. et al. MercuryII-mediated formation of thymine–HgII–thymine base pairs in DNA duplexes. J. Am. Chem. Soc. 128, 2172–2173 (2006).
Park, K. S., Jung, C. & Park, H. G. ‘Illusionary’ polymerase activity triggered by metal ions: use for molecular logic-gate operations. Angew. Chem. Int. Ed. 49, 9757–9760 (2010).
Ono, A. et al. Specific interactions between silver(I) ions and cytosine–cytosine pairs in DNA duplexes. Chem. Commun. 39, 4825–4827 (2008).
Urata, H., Yamaguchi, E., Nakamura, Y. & Wada, S-i. Pyrimidine–pyrimidine base pairs stabilized by silver(I) ions. Chem. Commun. 47, 941–943 (2011).
Carell, T. Molecular computing: DNA as a logic operator. Nature 469, 45–46 (2011).
Czlapinski, J. L. & Sheppard, T. L. Nucleic acid template-directed assembly of metallosalen–DNA conjugates. J. Am. Chem. Soc. 123, 8618–8619 (2001).
Czlapinski, J. L. & Sheppard, T. L. Template-directed assembly of metallosalen–DNA hairpin conjugates. ChemBioChem 5, 127–129 (2004).
Gothelf, K. V., Thomsen, A., Nielsen, M., Cló, E. & Brown, R. S. Modular DNA-programmed assembly of linear and branched conjugated nanostructures. J. Am. Chem. Soc. 126, 1044–1046 (2004).
Nielsen, M., Thomsen, A. H., Cló, E., Kirpekar, F. & Gothelf, K. V. Synthesis of linear and tripoidal oligo(phenylene ethynylene)-based building blocks for application in modular DNA-programmed assembly. J. Org. Chem. 69, 2240–2250 (2004).
Clever, G. H., Polborn, K. & Carell, T. A highly DNA-duplex-stabilizing metal-salen base pair. Angew. Chem. Int. Ed. 44, 7204–7208 (2005).
Clever, G. H. & Carell, T. Controlled stacking of 10 transition-metal ions inside a DNA duplex. Angew. Chem. Int. Ed. 46, 250–253 (2007).
Clever, G. H., Reitmeier, S. J., Carell, T. & Schiemann, O. Antiferromagnetic coupling of stacked cuII-salen complexes in DNA. Angew. Chem. Int. Ed. 49, 4927–4929 (2010).
Clever, G. H., Soeltl, Y., Burks, H., Spahl, W. & Carell, T. Metal–salen-base-pair complexes inside DNA: complexation overrides sequence information. Chem. Eur. J. 12, 8708–8718 (2006).
Clever, G. H., Kaul, C. & Carell, T. DNA–metal base pairs. Angew. Chem. Int. Ed. 46, 6226–6236 (2007).
Müller, J. Metal-ion-mediated base pairs in nucleic acids. Eur. J. Inorg. Chem. 3749–3763 (2008).
Clever, G. H. & Shionoya, M. Metal–base pairing in DNA. Coord. Chem. Rev. 254, 2391–2402 (2010).
Yoon, T. P. & Jacobsen, E. N. Privileged chiral catalysts. Science 299, 1691–1693 (2003).
Atwell, S., Meggers, E., Spraggon, G. & Schultz, P. G. Structure of a copper-mediated base pair in DNA. J. Am. Chem. Soc. 123, 12364–12367 (2001).
Johannsen, S., Megger, N., Boehme, D., Sigel, R. K. O. & Mueller, J. Solution structure of a DNA double helix with consecutive metal-mediated base pairs. Nature Chem. 2, 229–234 (2010).
Johannsen, S., Paulus, S., Düpre, N., Müller, J. & Sigel, R. K. O. Using in vitro transcription to construct scaffolds for one-dimensional arrays of mercuric ions. J. Inorg. Biochem. 102, 1141–1151 (2008).
Acknowledgements
The authors acknowledge financial support from the Deutsche Forschungsgemeinschaft (SFB749, CiPSM) and the Volkswagen Foundation (grant no. I/85052). C.K. is the recipient of a pre-doctoral fellowship from the Fonds der Chemischen Industrie.
Author information
Authors and Affiliations
Contributions
C.K. and M.W. performed the chemical experiments. C.K. carried out the biochemical assays and crystallization experiments. M.M. solved the crystal structures and supervised the biological studies. S.S. collected crystal data. T.C. designed the study and supervised the research project. C.K., M.M. and T.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary information
Supplementary information (PDF 1255 kb)
Rights and permissions
About this article
Cite this article
Kaul, C., Müller, M., Wagner, M. et al. Reversible bond formation enables the replication and amplification of a crosslinking salen complex as an orthogonal base pair. Nature Chem 3, 794–800 (2011). https://doi.org/10.1038/nchem.1117
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchem.1117
This article is cited by
-
Functionalization of acyclic xenonucleic acid with modified nucleobases
Polymer Journal (2023)
-
Stable Hg(II)-mediated base pairs with a phenanthroline-derived nucleobase surrogate in antiparallel-stranded DNA
JBIC Journal of Biological Inorganic Chemistry (2020)
-
Construction and characterization of metal ion-containing DNA nanowires for synthetic biology and nanotechnology
Scientific Reports (2019)
-
Silver(I)-mediated base pairing in parallel-stranded DNA involving the luminescent cytosine analog 1,3-diaza-2-oxophenoxazine
JBIC Journal of Biological Inorganic Chemistry (2019)
-
DNA nanotechnology
Nature Reviews Materials (2017)