Abstract
Stromal fibroblast senescence has been linked to ageing-associated cancer risk. However, density and proliferation of cancer-associated fibroblasts (CAFs) are frequently increased. Loss or downmodulation of the Notch effector CSL (also known as RBP-Jκ) in dermal fibroblasts is sufficient for CAF activation and ensuing keratinocyte-derived tumours. We report that CSL silencing induces senescence of primary fibroblasts from dermis, oral mucosa, breast and lung. CSL functions in these cells as a direct repressor of multiple senescence- and CAF-effector genes. It also physically interacts with p53, repressing its activity. CSL is downmodulated in stromal fibroblasts of premalignant skin actinic keratosis lesions and squamous cell carcinomas, whereas p53 expression and function are downmodulated only in the latter, with paracrine FGF signalling as the probable culprit. Concomitant loss of CSL and p53 overcomes fibroblast senescence, enhances expression of CAF effectors and promotes stromal and cancer cell expansion. The findings support a CAF activation–stromal co-evolution model under convergent CSL–p53 control.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Change history
28 August 2015
An error in the print version of this Article meant that Witold W. Kilarski's name was incorrect. This has been corrected in all online versions.
References
Bissell, M. J. & Hines, W. C. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 17, 320–329 (2011).
Gatenby, R. Perspective: finding cancer’s first principles. Nature 491, S55 (2012).
Martincorena, I. et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science 348, 880–886 (2015).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Campisi, J., Andersen, J. K., Kapahi, P. & Melov, S. Cellular senescence: a link between cancer and age-related degenerative disease? Semin. Cancer Biol. 21, 354–359 (2011).
Kuilman, T. & Peeper, D. S. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer 9, 81–94 (2009).
van Deursen, J. M. The role of senescent cells in ageing. Nature 509, 439–446 (2014).
Junttila, M. R. & de Sauvage, F. J. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 501, 346–354 (2013).
Polanska, U. M. & Orimo, A. Carcinoma-associated fibroblasts: non-neoplastic tumour-promoting mesenchymal cells. J. Cell Physiol. 228, 1651–1657 (2013).
Hoenicke, L. & Zender, L. Immune surveillance of senescent cells–biological significance in cancer- and non-cancer pathologies. Carcinogenesis 33, 1123–1126 (2012).
Bar, J., Moskovits, N. & Oren, M. Involvement of stromal p53 in tumor-stroma interactions. Semin. Cell Dev. Biol. 21, 47–54 (2010).
Polyak, K., Haviv, I. & Campbell, I. G. Co-evolution of tumor cells and their microenvironment. Trends Genet. 25, 30–38 (2009).
Tlsty, T. D. & Coussens, L. M. Tumor stroma and regulation of cancer development. Annu. Rev. Pathol. 1, 119–150 (2006).
Weinberg, R. A. Coevolution in the tumor microenvironment. Nat. Genet. 40, 494–495 (2008).
Coppe, J. P. et al. Tumor suppressor and aging biomarker p16(INK4a) induces cellular senescence without the associated inflammatory secretory phenotype. J. Biol. Chem. 286, 36396–36403 (2011).
Kopan, R. & Ilagan, M. X. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137, 216–233 (2009).
Hu, B. et al. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell 149, 1207–1220 (2012).
Ng, Y. Z. et al. Fibroblast-derived dermal matrix drives development of aggressive cutaneous squamous cell carcinoma in patients with recessive dystrophic epidermolysis bullosa. Cancer Res. 72, 3522–3534 (2012).
Costea, D. E. et al. Identification of two distinct carcinoma-associated fibroblast subtypes with differential tumor-promoting abilities in oral squamous cell carcinoma. Cancer Res. 73, 3888–3901 (2013).
Hosein, A. N. et al. Breast carcinoma-associated fibroblasts rarely contain p53 mutations or chromosomal aberrations. Cancer Res. 70, 5770–5777 (2010).
Navab, R. et al. Prognostic gene-expression signature of carcinoma-associated fibroblasts in non-small cell lung cancer. Proc. Natl Acad. Sci. USA 108, 7160–7165 (2011).
Iannello, A., Thompson, T. W., Ardolino, M., Lowe, S. W. & Raulet, D. H. p53-dependent chemokine production by senescent tumor cells supports NKG2D-dependent tumor elimination by natural killer cells. J. Exp. Med. 210, 2057–2069 (2013).
Novakova, Z. et al. Cytokine expression and signaling in drug-induced cellular senescence. Oncogene 29, 273–284 (2010).
Yoon, I. K. et al. Exploration of replicative senescence-associated genes in human dermal fibroblasts by cDNA microarray technology. Exp. Gerontol. 39, 1369–1378 (2004).
Freund, A., Patil, C. K. & Campisi, J. p38MAPK is a novel DNA damage response-independent regulator of the senescence-associated secretory phenotype. EMBO J. 30, 1536–1548 (2011).
Hinz, B. et al. Recent developments in myofibroblast biology: paradigms for connective tissue remodeling. Am. J. Pathol. 180, 1340–1355 (2012).
Riley, T., Sontag, E., Chen, P. & Levine, A. Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008).
Wienken, C. J., Baaske, P., Rothbauer, U., Braun, D. & Duhr, S. Protein-binding assays in biological liquids using microscale thermophoresis. Nat. commun. 1, 100 (2010).
Lill, N. L., Grossman, S. R., Ginsberg, D., DeCaprio, J. & Livingston, D. M. Binding and modulation of p53 by p300/CBP coactivators. Nature 387, 823–827 (1997).
Katoh, M. & Nakagama, H. FGF receptors: cancer biology and therapeutics. Med. Res. Rev. 34, 280–300 (2014).
Zhou, W. et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 17, 170–182 (2015).
Erez, N., Truitt, M., Olson, P., Arron, S. T. & Hanahan, D. Cancer-Associated Fibroblasts Are Activated in Incipient Neoplasia to Orchestrate Tumor-Promoting Inflammation in an NF-κB-Dependent Manner. Cancer Cell 17, 135–147 (2010).
Quante, M. et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell 19, 257–272 (2011).
Dotto, G. P. Multifocal epithelial tumors and field cancerization: stroma as a primary determinant. J. Clin. Invest. 124, 1446–1453 (2014).
Salem, A. F. et al. Cigarette smoke metabolically promotes cancer, via autophagy and premature aging in the host stromal microenvironment. Cell Cycle 12, 818–825 (2013).
Castel, D. et al. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 27, 1059–1071 (2013).
Johnson, J. E. & Macdonald, R. J. Notch-independent functions of CSL. Curr. Top. Dev. Biol. 97, 55–74 (2011).
Dotto, G. P. Crosstalk of Notch with p53 and p63 in cancer growth control. Nat. Rev. Cancer 9, 587–595 (2009).
Chene, P. Inhibition of the p53-MDM2 interaction: targeting a protein-protein interface. Mol. Cancer Res. 2, 20–28 (2004).
Del Bianco, C., Aster, J. C. & Blacklow, S. C. Mutational and energetic studies of Notch 1 transcription complexes. J. Mol. Biol. 376, 131–140 (2008).
Friedmann, D. R., Wilson, J. J. & Kovall, R. A. RAM-induced allostery facilitates assembly of a notch pathway active transcription complex. J. Biol. Chem. 283, 14781–14791 (2008).
Lujambio, A. et al. Non-cell-autonomous tumor suppression by p53. Cell 153, 449–460 (2013).
Cooks, T. et al. Mutant p53 prolongs NF-kappaB activation and promotes chronic inflammation and inflammation-associated colorectal cancer. Cancer cell 23, 634–646 (2013).
Devgan, V., Mammucari, C., Millar, S. E., Brisken, C. & Dotto, G. P. p21WAF1/Cip1 is a negative transcriptional regulator of Wnt4 expression downstream of Notch1 activation. Genes Dev. 19, 1485–1495 (2005).
Dotto, G. P. p21WAF1/Cip1: more than a break to the cell cycle? Biochim. Biophys. Acta 87483, 1–14 (2000).
Warfel, N. A. & El-Deiry, W. S. p21WAF1 and tumourigenesis: 20 years after. Curr. Opin. Oncol. 25, 52–58 (2013).
Sun, Y., Zeng, X. R., Wenger, L., Firestein, G. S. & Cheung, H. S. P53 down-regulates matrix metalloproteinase-1 by targeting the communications between AP-1 and the basal transcription complex. J. Cell. Biochem. 92, 258–269 (2004).
Boggs, K. & Reisman, D. C/EBPβ participates in regulating transcription of the p53 gene in response to mitogen stimulation. J. Biol. Chem. 282, 7982–7990 (2007).
Bruno, T. et al. Che-1 phosphorylation by ATM/ATR and Chk2 kinases activates p53 transcription and the G2/M checkpoint. Cancer Cell 10, 473–486 (2006).
Kolev, V. et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat. Cell Biol. 10, 902–911 (2008).
Phan, R. T. & Dalla-Favera, R. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature 432, 635–639 (2004).
Reisman, D. & Loging, W. T. Transcriptional regulation of the p53 tumor suppressor gene. Semin. Cancer Biol. 8, 317–324 (1998).
Rowland, B. D., Bernards, R. & Peeper, D. S. The KLF4 tumour suppressor is a transcriptional repressor of p53 that acts as a context-dependent oncogene. Nat. Cell Biol. 7, 1074–1082 (2005).
Shaulian, E. & Karin, M. AP-1 in cell proliferation and survival. Oncogene 20, 2390–2400 (2001).
Stepniak, E. et al. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 20, 2306–2314 (2006).
Malchers, F. et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer Discov. 4, 246–257 (2014).
Lefort, K. et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 21, 562–577 (2007).
Hu, B. et al. Control of hair follicle cell fate by underlying mesenchyme through a CSL-Wnt5a-FoxN1 regulatory axis. Genes Dev. 24, 1519–1532 (2010).
Lefort, K. et al. Notch1 is a p53 target gene involved in human keratinocyte tumor suppression through negative regulation of ROCK1/2 and MRCKalpha kinases. Genes Dev. 21, 562–577 (2007).
Restivo, G. et al. IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes. EMBO J. 30, 4571–4585 (2011).
Rheinwald, J. G. & Beckett, M. A. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 41, 1657–1663 (1981).
Gioanni, J. et al. Two new human tumor cell lines derived from squamous cell carcinomas of the tongue: establishment, characterization and response to cytotoxic treatment. Eur. J. Cancer Clin. Oncol. 24, 1445–1455 (1988).
Wu, X. et al. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature 465, 368–372 (2010).
el-Deiry, W. S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell 75, 817–825 (1993).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Wu, L. et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26, 484–489 (2000).
Lefort, K. et al. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 32, 2248–2263 (2013).
Kilarski, W. W. et al. Intravital immunofluorescence for visualizing the microcirculatory and immune microenvironments in the mouse ear dermis. PLoS ONE 8, e57135 (2013).
Smyth, G. K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, a3 (2004).
Acknowledgements
We thank R. Bernards, R. Agami, W. Raffoul, J. P. Rival, J. M. Joseph, U. Just, F. Martinon and T. Petrova for vectors or cells, F. De Sousa e Melo and B. Hu for early contributions, K. Harshman for RNA-Seq and ChIP-Seq analysis and C. Pasche and T. Proust for technical help. The work was supported by grants from the Swiss National Science Foundation (310030_156191/1), National Institutes of Health (R01AR039190; R01AR064786; the content not necessarily representing the official views of NIH), European Research Council (26075083) and OncoSuisse (OCS-2922-02-2012 and KFS-3301-08-2013). P. Ostano was supported by a grant from Lauretana S.P.A.
Author information
Authors and Affiliations
Contributions
M.-G.P., C.L., D.A.L., D.E.K., P.B., S.-H.J., S.G., E.M. and K.L. carried out work and contributed to analysis of results. P.O., U.A. and P.P. conducted bioinformatics. W.H. and V.N. provided clinical samples. W.W.K. and M.A.S. contributed to ear injection experiments. C.B. contributed experimental reagents and insights. G.P.D. designed the study. M.-G.P., K.L. and G.P.D. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Stromal cell senescence in mice with mesenchymal CSL (RBP-Jκ) deletion.
(a) Skin of a newborn (P0) mouse with deletion of the CSL (RBP-Jκ) gene (KO2), different from the one shown in Fig. 1a, was analyzed for SA-β-Gal activity in parallel with immunofluorescence with anti-vimentin antibodies for fibroblast identification. (b) Two additional skin lesions of 3 months old CSL mutant mice (male), besides that in Fig. 1b, were analyzed by double immunofluorescence with antibodies against keratin 14 (K14) and Tenascin C (upper panels) and, in parallel sections, for SA-β-Gal activity and anti-vimentin immunofluorescence (lower panels). (c) Parallel sections of the skin lesions analyzed above were stained with antibodies against vimentin F4/80 and phospho-histone 3 (pH3). Quantification of proliferating (phospho-histone 3 positive) fibroblasts (vimentin positive and F4/80 negative) is shown in Fig. 1c.
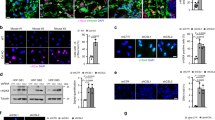
Supplementary Figure 2 CSL as negative regulator of dermal fibroblast senescence- and CAF-determinant genes.
(a) HDFs infected with two shRNA lentiviruses against CSL were analyzed, 3 days later, for CSL expression by immunoblotting. Densitometric quantification showed reduction of CSL to 3% and 6% of control. Unprocessed original scans of blots are shown in Supplementary Fig. 9. (b) SA-ß gal and clonogenicity assays of foreskin-derived HDFs (strain no. 4) plus/minus CSL silencing as shown in Fig. 1f, g for strain no. 1 adult-derived HDFs; for SA-β-Gal activity n = 290 (control shRNA), 264 (CSL1 shRNA), 217 (CSL2 shRNA) cells assessed from 4, 9 and 11 fields, respectively; mean ± s.d., two-tailed unpaired t-test, ∗P < 0.05. (c) Clonogenicity assays of another adult-derived HDF strain (no. 2) plus/minus CSL silencing; n = 3 biological replicates/condition, two-tailed unpaired t-test, ∗P < 0.05. (d) HDFs plus/minus CSL silencing were tested by 5-bromo-2‘-deoxyuridine (BrdU) incorporation assay; n = 113(control shRNA), 97 (CSL1 shRNA), 108 (shCSL2) cells assessed from 8 fields, mean ± s.d., two-sided one sample t-test, ∗P < 0.0005. (e) Cells as in (d) were tested for Alamar blue fluorescence cell density assays; data are presented as fluorescence intensity change over day 1. (f and g) Two HDF strains (no. 1 and no. 4) plus/minus CSL silencing by shRNAs (f) and siRNAs (g) examined for senescence-determinant genes in Fig. 1h, i were analyzed by RT-qPCR for indicated CAF marker genes. (h) Immunofluorescence analysis of HDFs plus/minus CSL silencing by siRNAs for p21WAF1/Cip1 and IL6, with DAPI staining for cell identification. Shown are representative images and quantification of fluorescence signal intensity by Image J software analysis. For p21 n = 164 (siCtrl), 190 (siCSL1), 114 (siCSL2) cells, for p53 and IL6 n = 222 (siCtrl), 194 (siCSL1), 97 (siCSL2) cells assessed from 4 fields. Whisker boxes: min to max, median center; two-tailed unpaired t-test, ∗P < 0.05. (i) Early passage oral (HOF), breast (HBF), lung (HLF) fibroblasts and WI38 diploid lung fibroblast line plus/minus shRNA-mediated CSL silencing as in Fig. 1k, l were tested by clonogenicity assays; for WI38 n = 3 biological replicates/condition, mean ± s.d., two-tailed unpaired t-test, ∗P < 0.05. (j) Primary human keratinocytes plus/minus siRNA-mediated CSL silencing were analyzed, 3 days after transfection, by RT-qPCR for indicated genes.
Supplementary Figure 3 Induction of fibroblast senescence and CAF marker-gene expression by increased Notch activity, and suppressive function of CSL.
(a) HDFs infected with a retrovirus expressing the cytoplasmic activated form of Notch1 fused to an ER domain (rNERT) in parallel with empty vector control (neo)43 were treated with 4-hydroxy-tamoxifen for 48 h to induce activation of the rNERT protein, followed by determination of SA-ß Gal activity; n = 78 (Neo) and 54 (NERT) cells assessed from 4 fields, mean ± s.d., two-tailed unpaired t-test, ∗P < 0.05. (b) HDFs infected with a lentivirus for doxycycline-inducible expression of the cytoplasmic activated form of Notch1 (pIND-N1) in parallel with empty vector control (pIND) were treated with doxycyclin for 5 days, followed by clonogenicity assay; n = 3 biological replicates/condition, mean ± s.d., two-tailed unpaired t-test, ∗P < 0.05. (c) HDFs as in the previous two panels were analyzed for expression of the indicated genes by RT-qPCR. (d) Human Oral Fibroblasts (HOF) stably infected with a lentivirus for doxycyclin-inducible expression of cytoplasmic Notch1 (pIND-N1), were treated for 48 h with doxycycline (+dox) or not (−dox) and analyzed for expression of indicated genes by RT-qPCR. (e) Two HDF strains (♯1 and ♯2) of independent origin were cultured for 48 h on dishes pre-coated with immunoglobulin-coupled Jagged 1 ligand or immunoglobulins alone (Ctrl), followed by RT-qPCR analysis of the indicated genes. (f) HDFs of two different strains (♯2 and ♯3), besides the one showed in Fig. 3a (♯1), were used for Chromatin Immuno-precipitation (ChIP) assays with two different antibodies against CSL in parallel with non-immune IgG controls, to assess binding to the indicated predicted CSL binding sites within each of the genes. (g) Graphic illustration of the position of CSL binding peaks revealed by ChIP-seq analysis with two different antibodies against CSL (red and blue colors) for the indicated genes, utilizing ENCODE information for promoter and enhancer localization, as indicated by islands of Histone H3 modifications (K4m3 and K27Ac, respectively), along with the respective position of Transcription Start Site (arrow) and coding exons (black boxes).
Supplementary Figure 4 Physical binding of the CSL and p53 proteins and CSL disruption of p53-p300 association.
(a and b) HDFs were immuno-precipitated with antibodies against CSL (a) or p53 (b) in parallel with non immune IgG, as in Fig. 4b, followed by immuno-blotting with antibodies against the CSL and p53 proteins as indicated. (c) Second independent MST analysis of purified recombinant CSL and p53 proteins incubated with each other as in Fig. 4e. Additionally, as positive control of high binding affinity, each of the purified recombinant CSL and p53 proteins was labeled with RED-NHS and admixed at a constant concentration (900 nM for p53, 250 nM for CSL) with two-fold serial dilutions of the corresponding specific antibodies (from 9 mM to 0.275 nM), followed by MST analysis. As negative control of specificity, purified recombinant p53 was admixed with two-fold serial dilutions of bovine serum albumin (from 9 mM to 0.275 nM), followed by MST analysis. Results are expressed as normalized thermophoresis-dependent fluorescence units (F-Norm) as a function of unlabeled ligand concentrations as in Fig. 4e. (d) Two independent experiments, besides what shown in Fig. 4i, with 293T cells co-transfected with a p53 plus/minus CSL expressing plasmids (3 μg of each in 10 cm dishes) followed, 24 h later, by immuno-precipitation of p300 and immuno-blotting with antibodies against p300 and p53. For (a–b) and (d) unprocessed original scans of blots are shown in Supplementary Fig. 9.
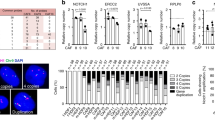
Supplementary Figure 5 Analysis of stromal cell senescence and gene expression in AKs, in situ and invasive skin SCCs.
(a–c) SA-β-Gal staining of two additional AKs (a), four in situ SCCs (b) and two invasive SCCs (c) besides those shown in Fig. 5a. (d) Larger fields and higher magnification images of in situ SCC (Fig. 5a) immune-fluorescence for vimentin and CD68 macrophage marker. CD68 positive cells were only deeply localized. (e) AKs of Fig. 5b (Patients 1-5) were analyzed by IF for vimentin and CD68. Representative image (P2 lesion) and quantification of vimentin-positive cells CD68-positive/negative; n(cells) = 130, 170, 155, 950, 150 (P1-P5) assessed from 3(P1-P2), 2(P3, P5) and 4(P4) fields. (f) RNA samples from 3rd passage HDFs, human THP1 monocytes after TPA-induced macrophage differentiation (MΦ) plus/minus LPS-triggered activation, and HUVEC endothelial cells, were analyzed individually and admixed in indicated amounts by RT-qPCR. (g) RNA samples from 2nd passage mouse dermal fibroblasts, primary mouse macrophages (MΦ) and endothelial cells (endo) were analyzed individually and admixed in indicated amounts by RT-qPCR. (h and i) Same LCM/RNA samples of Fig. 5c and e-g were examined by RT-qPCR for leukocyte/macrophage CD45 marker. (h) n = 5 AKs and 5 matched NS regions, mean ± s.e.m., two-tailed paired t-test; n = 5 SCC and 5 NS regions from different individuals, mean ± s.e.m., two-tailed unpaired t-test; n = 6 SCC regions (two per SCC) and 3 matched NS, mean ± s.e.m., two-tailed paired t-test; (i) n = 5 affected and 3 unaffected regions, ratio (affected/unaffected), two-sided one sample t-test. No signal was obtained for endothelial CD31 marker. (j) Representative images of in situ SCCs and normal skin sections processed for fluorescence-guided LCM utilizing anti-PDGRα-FITC-conjugated antibodies and Propidium iodide (PI) staining for nuclei identification. Shown are stained sections, selected cells before LCM (by InfraRed and UV cutting), corresponding empty spaces and captured material on LCM cap. (k) Quantification of CSL immunofluorescence signal in vimentin-positive and CD68-negative cells in stroma underlying three skin SCCs versus normal skin from different individuals and representative images of triple immuno-fluorescence analysis; n(vimentin-positive/CD68-negative cells) = 174, 178, 152 (NS) and 780, 488, 720 (AK), mean of signal intensity ± s.e.m., two tailed unpaired t-test.
Supplementary Figure 6 Impact of various growth factors and TK inhibitors on fibroblast p53 gene expression and activity.
(a) HDFs of independent origin from those tested in Fig. 6b were similarly treated with the indicated growth factors/cytokines (at the concentrations specified in online Methods) followed, 72 h later, by RT-qPCR analysis of p53 expression. (b and c) HDFs of a different strain from that tested in Fig. 6d were similarly treated with the Ponatinib or Imatinib inhibitors at the indicated concentrations followed, 72 h later, by RT-qPCR analysis of p53, CDKN1A and miR34a expression (b) or p53 immunoblotting (c). Unprocessed original scans of blots are shown in Supplementary Fig. 9. (d) HDFs cultured in the presence or absence of Ponatinib (100 nM) or Imatinib (5 μM) for 7 days were assayed for SA-ß-gal activity or clonogenicity in triplicate dishes. For SA-ß-gal n = 771 (DMSO), 200 (Ponatinib), 337 (Imatinib) cells assessed from 4 fields; for clonogenicity assay n = 3 biological replicates/condition, mean ± s.d.. Shown are representative images of cells after SA-ß-gal staining as well as quantification of the data (left and right panels, respectively). (e) Similar CAF cultures as in Fig. 6f, g (strain no. 1) were cultured in the presence or absence of Ponatinib or Imatinib followed by assay for SA-ß-gal activity; n = 240 (DMSO), 105 (Ponatinib 100 nM), 67 (Ponatinib 500 nM), 124 (Imatinib) cells assessed from 3, 10, 7 and 8 fields, respectively. Shown is mean ± s.d.
Supplementary Figure 7 Abrogation of HDF senescence with enhancement of CAF marker expression by concomitant CSL and p53 silencing or gene disruption.
(a) Early passage HDFs were infected with a CRISPR lentivirus against p53, followed by selection for the virally-transduced antibiotic resistance gene (puromycin). Resulting colonies (>500) were pooled and analyzed (CRISPR), in parallel with parental control cells (WT), for p53 protein levels by immuno-blotting, and for expression of p53 target genes (p21WAF1/Cip1, miR34-a, MDM2) by RT-qPCR (left and right panels, respectively). Similar results were obtained with two other strain of HDFs of independent origin. Unprocessed original scans of blots are shown in Supplementary Fig. 9. (b) HDFs (a second strain besides that tested in Fig. 7a) were infected with control versus p53 silencing retroviruses, followed by infection with control versus CSL silencing lentiviruses. Number of cells positive for SA-ß-Gal activity was determined a week later (left panel); from left to right n (cells) = 372, 223, 236, 332, 370, 373 assessed from 4, 6, 8, 4, 9 and 4 fields, respectively. The same cells were also plated under sparse conditions, followed by determination of colony formation 10 days later (right panel); n = 3 biological replicates/condition. (c) Parallel cultures of cells as in the previous panel were analyzed by RT-qPCR for expression of the indicated genes. (d) HDFs were infected with control versus CDKN1A silencing retroviruses, followed by infection with either control or CSL silencing lentiviruses and testing by clonogenicity assays as in other experiments; n = 3 biological replicates/condition. (e) HDFs were transfected with siRNAs against CSL with or without siRNAs for CDKN1A, followed, 3 days later, by a repeated transfection. SA-ß-Gal activity was assessed at day seven; from left to right, n(cells) = 130, 111, 52, 223, 210, 106 assessed from 4, 6, 8, 4, 4, and 9 fields, respectively. (f) Parallel cultures of cells as in the previous panel were analyzed by RT-qPCR for expression of the indicated genes, with 36 β4 for normalization. For (b) and (d–e) mean ± s.d., two-tailed unpaired t-test is shown; ∗P < 0.05.
Supplementary Figure 8 Tumor and stromal cell expansion as a function of CSL and p53 stromal expression.
(a) DsRed2 expressing SCC13 cells were admixed with GFP expressing HDFs with shRNA-mediated silencing of CSL or p53 individually or in combination, followed by parallel injections into mouse ears and imaging every 2-3 days under a fluorescence dissection microscope as in Fig. 8a. Shown are images from another pair of mouse ear injections of SCC13 cells admixed with HDFs with combined CSL and p53 silencing versus CSL silencing alone, at the indicated times after injection. (b) Immuno-histochemical analysis of the indicated markers in lesions from another pair of mouse ear injections, besides that shown in Fig. 8d, of SCC13 cells admixed with HDFs with combined silencing of CSL and p53 versus CSL alone. (c) Left Panel: Head/Neck Cal27 SCC cells expressing DsRed2 were admixed with PKH67-green labeled HDFs with CSL silenced individually or in combination with p53, followed by parallel injections into mouse ears and imaging every 2–3 days under a fluorescence dissection microscope. Shown are representative images from one pair of mouse ears at the beginning and end of the experiment (24 days after injection). Right panel: Quantification of digital images for red (SCC cells) fluorescence intensity values (intensity x surface area) for the two combination of cells throughout the experiment with 3 NOD/SCID Il2rg−/− 10-weeks-old male mice/ear pairs per condition. To take into account individual animal variations, for each mouse pair, the signal increase in the ear injected with control cells was set to 1. Quantification was done by software (ImageJ) analysis of the digitally acquired images; n = 3 lesions per condition, mean ± s.e.m., one-tailed paired t-test at day 24, ∗P > 0.05 (d) Histological images of one pair of mouse ears at the end of the experiment at low (upper) and high (lower) magnification. (e) Double immunofluorescence analysis with anti-vimentin and phospho-Histone 3 antibodies, showing increased proliferation in both vimentin negative and positive cells.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1210 kb)
Supplementary Information
Supplementary Table 1 (XLSX 319 kb)
Supplementary Information
Supplementary Table 2 (XLSX 2964 kb)
Supplementary Information
Supplementary Table 3 (XLSX 39 kb)
Supplementary Information
Supplementary Table 4 (XLSX 52 kb)
Rights and permissions
About this article
Cite this article
Procopio, MG., Laszlo, C., Al Labban, D. et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol 17, 1193–1204 (2015). https://doi.org/10.1038/ncb3228
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3228
This article is cited by
-
Aging microenvironment and antitumor immunity for geriatric oncology: the landscape and future implications
Journal of Hematology & Oncology (2023)
-
Heterogeneity of cancer-associated fibroblasts in head and neck squamous cell carcinoma: opportunities and challenges
Cell Death Discovery (2023)
-
Whole exome and transcriptome analysis revealed the activation of ERK and Akt signaling pathway in canine histiocytic sarcoma
Scientific Reports (2023)
-
The roles and mechanisms of senescence-associated secretory phenotype (SASP): can it be controlled by senolysis?
Inflammation and Regeneration (2022)
-
Enterobacteria impair host p53 tumor suppressor activity through mRNA destabilization
Oncogene (2022)