Abstract
Focal adhesions (FAs) link the extracellular matrix to the actin cytoskeleton to mediate cell adhesion, migration, mechanosensing and signalling. FAs have conserved nanoscale protein organization, suggesting that the position of proteins within FAs regulates their activity and function. Vinculin binds different FA proteins to mediate distinct cellular functions, but how vinculin’s interactions are spatiotemporally organized within FAs is unknown. Using interferometric photoactivation localization super-resolution microscopy to assay vinculin nanoscale localization and a FRET biosensor to assay vinculin conformation, we found that upward repositioning within the FA during FA maturation facilitates vinculin activation and mechanical reinforcement of FAs. Inactive vinculin localizes to the lower integrin signalling layer in FAs by binding to phospho-paxillin. Talin binding activates vinculin and targets active vinculin higher in FAs where vinculin can engage retrograde actin flow. Thus, specific protein interactions are spatially segregated within FAs at the nanoscale to regulate vinculin activation and function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Kuo, J-C. C., Han, X., Hsiao, C-T. T., Yates Iii, J. R. & Waterman, C. M. Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13, 383–393 (2011).
Byron, A., Humphries, J. D., Bass, M. D., Knight, D. & Humphries, M. J. Proteomic analysis of integrin adhesion complexes. Sci. Signal. 4, pt2 (2011).
Schiller, H. B., Friedel, C. C., Boulegue, C. & Fässler, R. Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep. 12, 259–266 (2011).
Kanchanawong, P. et al. Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 (2010).
Choi, C. K. et al. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 (2008).
Zaidel-Bar, R., Ballestrem, C., Kam, Z. & Geiger, B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116, 4605–4613 (2003).
Zaidel-Bar, R., Milo, R., Kam, Z. & Geiger, B. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120, 137–148 (2007).
Zamir, E. et al. Molecular diversity of cell-matrix adhesions. J. Cell Sci. 112, 1655–1669 (1999).
Wolfenson, H. et al. A role for the juxtamembrane cytoplasm in the molecular dynamics of focal adhesions. PLoS ONE 4, e4304 (2009).
Diez, G., Auernheimer, V., Fabry, B. & Goldmann, W. H. Head/tail interaction of vinculin influences cell mechanical behavior. Biochem. Biophys. Res. Commun. 406, 85–88 (2011).
Dumbauld, D. W. et al. How vinculin regulates force transmission. Proc. Natl Acad. Sci. USA 110, 9788–9793 (2013).
Saunders, R. M. et al. Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 85, 487–500 (2006).
Humphries, J. D. et al. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J. Cell Biol. 179, 1043–1057 (2007).
Carisey, A. et al. Vinculin regulates the recruitment and release of core focal adhesion proteins in a force-dependent manner. Curr. Biol. 23, 271–281 (2013).
Plotnikov, S. V., Pasapera, A. M., Sabass, B. & Waterman, C. M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 (2012).
Thievessen, I. et al. Vinculin-actin interaction couples actin retrograde flow to focal adhesions, but is dispensable for focal adhesion growth. J. Cell Biol. 202, 163–177 (2013).
Subauste, M. C. et al. Vinculin modulation of paxillin-FAK interactions regulates ERK to control survival and motility. J. Cell Biol. 165, 371–381 (2004).
Burridge, K. & Mangeat, P. An interaction between vinculin and talin. Nature 308, 744–746 (1984).
Jockusch, B. M. & Isenberg, G. Interaction of α-actinin and vinculin with actin: opposite effects on filament network formation. Proc. Natl Acad. Sci. USA 78, 3005–3009 (1981).
Turner, C. E., Glenney, J. R. & Burridge, K. Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 111, 1059–1068 (1990).
Gilmore, A. P. & Burridge, K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4-5-bisphosphate. Nature 381, 531–535 (1996).
DeMali, K. A., Barlow, C. A. & Burridge, K. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J. Cell Biol. 159, 881–891 (2002).
Kioka, N. et al. Vinexin: a novel vinculin-binding protein with multiple SH3 domains enhances actin cytoskeletal organization. J. Cell Biol. 144, 59–69 (1999).
Johnson, R. P. & Craig, S. W. An intramolecular association between the head and tail domains of vinculin modulates talin binding. J. Biol. Chem. 269, 12611–12619 (1994).
Johnson, R. P. & Craig, S. W. F-actin binding site masked by the intramolecular association of vinculin head and tail domains. Nature 373, 261–264 (1995).
Chen, H., Choudhury, D. M. & Craig, S. W. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 281, 40389–40398 (2006).
Calderwood, D. A. et al. The talin head domain binds to integrin subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. 274, 28071–28074 (1999).
Goldmann, W. H. et al. Examining F-actin interaction with intact talin and talin head and tail fragment using static and dynamic light scattering. Eur. J. Biochem. 250, 447–450 (1997).
Critchley, D. R. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. 38, 235–254 (2009).
Rubashkin, M. G. et al. Force engages vinculin and promotes tumor progression by enhancing PI3K activation of phosphatidylinositol (3,4,5)-triphosphate. Cancer Res. 74, 4597–4611 (2014).
Bakolitsa, C. et al. Structural basis for vinculin activation at sites of cell adhesion. Nature 430, 583–586 (2004).
Cohen, D. M., Chen, H., Johnson, R. P., Choudhury, B. & Craig, S. W. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 280, 17109–17117 (2005).
Cohen, D. M., Kutscher, B., Chen, H., Murphy, D. B. & Craig, S. W. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 281, 16006–16015 (2006).
Ciobanasu, C., Faivre, B. & Le Clainche, C. Actomyosin-dependent formation of the mechanosensitive talin-vinculin complex reinforces actin anchoring. Nat. Commun. 5, 3095 (2014).
Del Rio, A. et al. Stretching single talin rod molecules activates vinculin binding. Science 323, 638–641 (2009).
Chen, H., Cohen, D. M., Choudhury, D. M., Kioka, N. & Craig, S. W. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 169, 459–470 (2005).
Shaner, N. C. et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013).
Pasapera, A. M., Schneider, I. C., Rericha, E., Schlaepfer, D. D. & Waterman, C. M. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188, 877–890 (2010).
Brown, M. C., Perrotta, J. A. & Turner, C. E. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J. Cell Biol. 135, 1109–1023 (1996).
Thompson, P. M. et al. Identification of an actin binding surface on vinculin that mediates mechanical cell and focal adhesion properties. Structure 22, 697–706 (2014).
Gardel, M. L. et al. Traction stress in focal adhesions correlates biphasically with actin retrograde flow speed. J. Cell Biol. 183, 999–1005 (2008).
Bachir, A. I. et al. Integrin-associated complexes form hierarchically with variable stoichiometry in nascent adhesions. Curr. Biol. 24, 1845–1853 (2014).
Wolfenson, H., Bershadsky, A., Henis, Y. I. & Geiger, B. Actomyosin-generated tension controls the molecular kinetics of focal adhesions. J. Cell Sci. 124, 1425–1432 (2011).
Schneider, I. C., Hays, C. K. & Waterman, C. M. Epidermal growth factor-induced contraction regulates paxillin phosphorylation to temporally separate traction generation from de-adhesion. Mol. Biol. Cell 20, 3155–3167 (2009).
Tadokoro, S. et al. Talin binding to integrin β tails: a final common step in integrin activation. Science 302, 103–106 (2003).
Astrof, N. S., Salas, A., Shimaoka, M., Chen, J. & Springer, T. A. Importance of force linkage in mechanochemistry of adhesion receptors. Biochemistry 45, 15020–15028 (2006).
Jiang, G., Giannone, G., Critchley, D. R., Fukumoto, E. & Sheetz, M. P. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature 424, 334–337 (2003).
Friedland, J. C., Lee, M. H. & Boettiger, D. Mechanically activated integrin switch controls α5β1 function. Science 323, 642–644 (2009).
Yao, M. et al. Mechanical activation of vinculin binding to talin locks talin in an unfolded conformation. Sci. Rep. 4, 4610 (2014).
Hoffmann, J-E., Fermin, Y., Stricker, R. L., Ickstadt, K. & Zamir, E. Symmetric exchange of multi-protein building blocks between stationary focal adhesions and the cytosol. eLife 3, e02257–e02257 (2014).
Shin, W. et al. in Live Cell Imaging: A Laboratory Manual (eds Goldman, R., Swedlow, J. & Spector, D.) 2nd edn (Cold Spring Harbor Press, 2010).
Thévenaz, P., Ruttimann, U. E. & Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 7, 27–41 (1998).
Shtengel, G. et al. Imaging cellular ultrastructure by PALM, iPALM, and correlative iPALM-EM. Methods Cell Biol. 123, 273–294 (2014).
Acknowledgements
The authors thank S. Craig (Johns Hopkins University) for the cDNA encoding the vinculin FRET activation biosensor, G. Danuser (UT Southwestern) for the FRET biosensor image analysis package, W. Shin for maintenance of the Waterman Lab microscopes, D. Honemond and S. Thacker for administrative assistance, T. Kanchanawong (National University of Singapore) for sharing and discussing iPALM protocols, and members of the Waterman Lab, G. Alushin (NHLBI), H. Elliot (Harvard Medical School) and J. Taraska (NHLBI) for helpful discussions. Financial support: Division of Intramural Research, NHLBI (L.B.C. and C.M.W.); Howard Hughes Medical Institute (G.S. and H.F.H.); GM081764 (S.L.C.), GM080568 (S.L.C.).
Author information
Authors and Affiliations
Contributions
L.B.C. and C.M.W. conceived the study and wrote the manuscript with input from all authors. L.B.C., C.M.W. and S.L.C. designed experiments. L.B.C. performed and analysed most experiments. L.B.C. and G.S. performed iPALM imaging. M.A.B. and M.W.D. designed new cDNA constructs and performed cloning. G.S. and H.F.H. conceived of, built and maintained iPALM instrumentation and developed iPALM processing tools. L.B.C., M.A.B., M.W.D. and C.M.W. designed the summary cartoon.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 4 Vinculin WT iPALM measurements do not significantly vary between experiments.
(a) Mean of Z-median measurements from individual FAs in cells expressing WT vinculin-N-tdEos and imaged in three independent experiments. (b) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence about the mean. Significance tested with two-sample KS test. (c) Mean fraction of molecules localized to each of the three FA layers in FAs. Colouring in b,c used to highlight the three FA layers as in Fig. 1. Graphs in a–c represent measurements of n = 21 FAs for 2 cells (Day1), n = 60 FAs from 3 cells (Day2), and n = 35 FAs from 3 cells (Day3). Data in all bar graphs are represented as mean ± 95% bootstrapped confidence intervals. Significance tested with one way ANOVA followed by post-hoc Tukey test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ∗∗∗∗∗p < 0.00001, ns: not significant.
Supplementary Figure 5 Talin nano-scale position is rescued by re-expression of WT Vinculin.
(a) Western blots of vinculin (top) and tubulin (loading control, bottom) protein in lysates of WT HFFs versus HFFs expressing vinculin siRNA for 72 h (KD). (b) Representative iPALM renderings from wild-type (WT) HFF cells expressing Talin-C-tdEos. (c,d) Representative iPALM rendering from HFF cells treated with siRNAs targeting vinculin (Vcl KD) and additionally expressing Talin-C-tdEos (c) or Talin-C-tdEos with Vinculin-mCerulean (c). (a,c data duplicated from Fig. 2 for comparison purposes). In b–d the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, and scale bar = 1 micron. Histograms of the Z-position of the molecules within individual FAs (white boxes in b–d) displayed next to the colourscale. (e) Mean of Z-median measurements of the position of molecules from individual FAs in cells expressing Talin-C-tdEos (TlnC) in WT HFFs (WT), TlnC in vinculin KD HFFs (Vcl KD) or in cells expressing TlnC and vinculin-mCerulean in vinculin KD HFFs (Res). (f,g) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence about the mean. Significance tested with two-sample KS test. (h,i) Mean fraction of molecules localized to each of the three FA layers in FAs from cells in e. Colouring in f–i used to highlight the three FA layers as in Fig. 1. Graphs in e–i represent measurements of n = 95 FAs from 5 TlnC WT cells, n = 66 FAs from 5 TlnC KD cells, and n = 79 FAs from 5 TlnC rescue cells. Data in all bar graphs are represented as mean ± 95% bootstrapped confidence intervals with significance tested with one way ANOVA followed by post-hoc Tukey test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ∗∗∗∗∗p < 0.00001, ns: not significant.
Supplementary Figure 6 Active vinculin is oriented with the tail above the head.
(a,b) Representative iPALM rendering from an HFF cell expressing N-terminally tagged wild-type constitutively active (CA, N773/E775A) vinculin-N-tdEos (a, data duplicated from Fig. 3 for comparison purposes), or C-terminally tagged CA-vinculin-C-tdEos (b). In a,b the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, and scale bar = 1 micron. Histograms of the Z-position of the molecules within individual FAs (white boxes in a,b) displayed next to the colourscale. (c) Mean of Z-median measurements from individual FAs. (d) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence about the mean. Significance tested with two-sample KS test. (e) Mean fraction of molecules localized to each of the three FA layers. Colouring in d,e used to highlight the three FA layers as in Fig. 1. Graphs in c–e represent measurements of n = 82 FAs from 8 CA-vinculin-N-tdEos cells and n = 148 FAs from 6 CA-vinculin-C-tdEos expressing cells. Data in all bar graphs are represented as mean ± 95% bootstrapped confidence intervals with significance tested with one way ANOVA. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ∗∗∗∗∗p < 0.00001, ns: not significant.
Supplementary Figure 7 Characterization of and controls for the vinculin activation FRET biosensor.
(a) mTurquoise (mTurq, top row) NeonGreen (NeonGr, middle row) and processed FRET ratio image (bottom row) of HFF cells either expressing mTurqouise fused to NeonGreen by a 10 amino acid linker (Cytosolic control probe), expressing the first 400 amino acids of vinculin (Vcl) fused to mTurquoise and NeonGreen (FA-targeted control probe), co-expressing mTurquoise and NeonGreen, or co-expressing vinculin-mTurquoise and vinculin-NeonGreen. Cartoon schematics of the different FRET probes are displayed beneath the images. Scale bar = 5 micron. In a,d, The FA mask (grey lines) was created from the mTurq images and superimposed onto FRET ratio images. (b) Quantification of the mean FRET ratio value for the constructs described in a in regions confined within FAs (FA) or in non-FA cytosolic regions (cyto). N = number of cells. (c) Scatterplot of the cytoplasmic mTurquoise Intensity versus Cytoplasmic FRET ratio measured in cells expressing FA-targeted control probe, WT-Vinculin FRET probe, co-expressing mTurquoise and NeonGreen, or co-expressing vinculin-mTurquoise and vinculin-NeonGreen. Each point represents measurements from a single cell. (d) mTurquoise (mTurq, top row) and processed FRET ratio image (middle row) of HFF cells expressing wild type (WT) vinculin FRET biosensor (left) or constitutively active (CA, N773/E775A) vinculin FRET biosensor (right). Scale bar = 5 micron. Bottom row: cartoon schematic of the WT vinculin FRET biosensor, numbers refer to amino acid positions in full-length vinculin. (d) Quantification of the mean FRET ratio value inside FAs (FA) and outside FAs (Cyto) from n = 18 WT-vinculin FRET expressing cells and n = 16 CA-vinculin FRET expressing cells. Data in all bar graphs are represented as mean ± 95% confidence intervals with significance tested with ANOVA test (∗∗∗∗∗p < 0.00001) followed by Tukey test post-hoc analysis. (∗ difference is significant at p < 0.05 cutoff, ns: not significant).
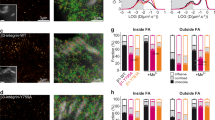
Supplementary Figure 8 Validation of paxillin knockdown and immunoprecipitation experiments, and the effects of paxillin overexpression on vinculin nanoscale localization.
(a) Western blots of Paxillin (top), Hic-5 (middle) or tubulin (loading control, bottom) protein in lysates of WT HFFs (WT) versus HFFs coexpressing paxillin and Hic-5 siRNA for 48 h (KD). (b) Western blots from Fig. 5d with molecular size markers labelled. (c) Independent co-immunoprecipitation displayed as a replicate experiment of Fig. 5d and Supplementary Fig. 5b. In b,c samples are from HFFs expressing vinculin siRNA for 72 hrs and additionally expressing GPF (1), WT-Vinculin-GFP (2), CA(N773/E775A)-Vinculin-GFP (3), A50I-Vinculin-GFP (4), or CA-A50I-Vinculin-GFP (5). (d,e) Representative iPALM renderings from HFF cells expressing WT-vinculin-tdEos (Vcl) and WT-paxillin-mCerulean (d) or WT-vinculin-tdEos (Vcl) and WT-paxillin-mCerulean in a paxillin/hic5 siRNA (Pxn KD) background (e). In d,e the colourscale represents Z-position (nm), FAs oriented with the distal tip facing up, scale bar = 1 micron. Histograms of the Z-position of the molecules within individual FAs (white boxes in d,e) are displayed next to the colourscale. (f) Mean of Z-median measurements of molecules in individual FAs. (g) Averaged Z-position frequency histograms of molecules within FAs. Solid line, mean frequency; Shaded region, bootstrapped 95% confidence about the mean. Significance tested with two-sample KS test. (h) Mean fraction of molecules localized to each of the three FA layers. Colouring in g,h used to highlight the three FA layers as in Fig. 1. Graphs in f–h represent measurements of n = 110 FA from 5 WT cells coexpressing WT-vinculin-N-tdEos and WT-paxillin-mCerulean and n = 111 FA from 5 PxnKD cells coexpressing WT-vinculin-N-tdEos and WT-paxillin-mCerulean. Data in all bar graphs represented as mean ± 95% bootstrapped confidence intervals with significance tested by one-way ANOVA.
Supplementary Figure 9 Spatial characterization of FRET biosensors and temporal characterization of paxillin phosphorylation during myosin-II dependent FA maturation.
(a,d) mTurquoise (mTurq, top) and processed mTurquoise/NeonGreen FRET ratio images (FRET, bottom) of a protruding region of an HFF cell expressing the CA(N773/E775A)-vinculin FRET biosensor (a) or the first 400 amino acids of vinculin (Vcl) fused to mTurquoise and NeonGreen (FA-targeted control probe, d). The FA mask (grey lines) was created from the mTurq image and superimposed onto FRET ratio image. Scale bar = 5 microns. (b,e) Values for mTurquoise intensity (blue line) and FRET ratio (red line) along the 5 μm long yellow line in a,d. Dark grey shaded areas are in the cytosolic (cyto) regions adjacent to the FA (FA, light grey shaded area) (c,f) Quantification of the mean FRET ratio of the distal (Dist) 1/3 and proximal (Prox) 1/3 of FA linescans. Data represented as mean ± standard error of measurements of n = 19 FA from 5 CA-vinculin FRET cells (c) or n = 18 FA from 7 control probe cells (f). Significance tested with one way ANOVA. In c,f, ‘Dist’ refers to the end of the FA facing the leading edge, and ‘Prox’ refers to the end of the FA facing the cell centre. (c) Uncropped western blots from Fig. 8r with molecular size markers labelled.
Supplementary Figure 10 Model of vinculin activation and nanoscale positioning during FA formation and maturation.
Speculative model for the role of vinculin-protein interactions in vinculin nanoscale localization and function during focal adhesion maturation. See text for details.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1236 kb)
Rights and permissions
About this article
Cite this article
Case, L., Baird, M., Shtengel, G. et al. Molecular mechanism of vinculin activation and nanoscale spatial organization in focal adhesions. Nat Cell Biol 17, 880–892 (2015). https://doi.org/10.1038/ncb3180
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3180
This article is cited by
-
pH-regulated single cell migration
Pflügers Archiv - European Journal of Physiology (2024)
-
Blockade of TGF-β signalling alleviates human adipose stem cell senescence induced by native ECM in obesity visceral white adipose tissue
Stem Cell Research & Therapy (2023)
-
Organization, dynamics and mechanoregulation of integrin-mediated cell–ECM adhesions
Nature Reviews Molecular Cell Biology (2023)
-
Remodeling of the focal adhesion complex by hydrogen-peroxide-induced senescence
Scientific Reports (2023)
-
Cellular nanomechanics derived from pattern-dependent focal adhesion and cytoskeleton to balance gene transfection of malignant osteosarcoma
Journal of Nanobiotechnology (2022)