Abstract
It has been postulated that there is a link between inflammation and cancer. Here we describe a role for cell-intrinsic toll-like receptor-2 (TLR2; which is involved in inflammatory response) signalling in normal intestinal and mammary epithelial cells and oncogenesis. The downstream effectors of TLR2 are expressed by normal intestinal and mammary epithelia, including the stem/progenitor cells. Deletion of MYD88 or TLR2 in the intestinal epithelium markedly reduces DSS-induced colitis regeneration and spontaneous tumour development in mice. Limiting dilution transplantations of breast epithelial cells devoid of TLR2 or MYD88 revealed a significant decrease in mammary repopulating unit frequency compared with the control. Inhibition of TLR2, its co-receptor CD14, or its downstream targets MYD88 and IRAK1 inhibits growth of human breast cancers in vitro and in vivo. These results suggest that inhibitors of the TLR2 pathway merit investigation as possible therapeutic and chemoprevention agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Metcalfe, C., Kljavin, N. M., Ybarra, R. & de Sauvage, F. J. Stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell 14, 149–159 (2014).
Yu, L., Wang, L. & Chen, S. Endogenous toll-like receptor ligands and their biological significance. J. Cell. Mol. Med. 14, 2592–2603 (2010).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
West, X. Z. et al. Oxidative stress induces angiogenesis by activating TLR2 with novel endogenous ligands. Nature 467, 972–976 (2010).
Park, J. S. High mobility group box 1 protein interacts with multiple Toll-like receptors. Am. J. Physiol. Cell Physiol. 290, C917–C924 (2005).
Kim, S. et al. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature 457, 102–106 (2009).
Yang, R-B. et al. Toll-like receptor-2 mediates lipopolysaccharide-induced cellular signalling. Nature 395, 284–288 (1998).
Lee, M. S. & Kim, Y-J. Signaling pathways downstream of pattern-recognition receptors and their cross talk. Annu. Rev. Biochem. 76, 447–480 (2007).
Wright, S. D., Ramos, R. A., Tobias, P. S., Ulevitch, R. J. & Mathison, J. C. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249, 1431–1433 (1990).
Clevers, H. At the crossroads of inflammation and cancer. Cell 118, 671–674 (2004).
Rakoff-Nahoum, S. & Medzhitov, R. Toll-like receptors and cancer. Nat. Rev. Cancer 9, 57–63 (2009).
Coussens, L. M. & Werb, Z. Inflammation and cancer. Nature 420, 860–867 (2002).
Balkwill, F. & Mantovani, A. Inflammation and cancer: back to Virchow? Lancet 357, 539–545 (2001).
Rakoff-Nahoum, S. & Medzhitov, R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science 317, 124–127 (2007).
Takeda, K., Kaisho, T. & Akira, S. Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 (2003).
Ngo, V. N. et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 470, 115–119 (2011).
Puente, X. S. et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 475, 101–105 (2011).
Pietersen, A. M. et al. Bmi1 regulates stem cells and proliferation and differentiation of committed cells in mammary epithelium. Curr. Biol. 18, 1094–1099 (2008).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Uemura, N. et al. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345, 784–789 (2001).
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013).
Medzhitov, R., Preston-Hurlburt, P. & Janeway, C. A. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388, 394–397 (1997).
Asselin-Labat, M-L. et al. Gata-3 negatively regulates the tumor-initiating capacity of mammary luminal progenitor cells and targets the putative tumor suppressor Caspase-14. Mol. Cell Biol. 31, 4609–4622 (2011).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006).
Stingl, J. et al. Purification and unique properties of mammary epithelial stem cells. Nature 439, 993–997 (2006).
Kordon, E. C. & Smith, G. H. An entire functional mammary gland may comprise the progeny from a single cell. Development 125, 1921–1930 (1998).
Schneider, D. S., Hudson, K. L., Lin, T. Y. & Anderson, K. V. Dominant and recessive mutations define functional domains of Toll, a transmembrane protein required for dorsal-ventral polarity in the Drosophila embryo. Genes Dev. 5, 797–807 (1991).
Keymeulen, A. V. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193 (2011).
Rios, A. C., Fu, N. Y., Lindeman, G. J. & Visvader, J. E. In situ identification of bipotent stem cells in the mammary gland. Nature 506, 322–327 (2014).
Plaks, V. et al. Lgr5-expressing cells are sufficient and necessary for postnatal mammary gland organogenesis. Cell Rep. 3, 70–78 (2013).
Grivennikov, S. I. et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258 (2012).
Sangiorgi, E. & Capecchi, M. R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40, 915–920 (2008).
Tian, H. et al. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature 478, 255–259 (2011).
Montgomery, R. K. et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc. Natl Acad. Sci. USA 108, 179–184 (2011).
Takeda, N. et al. Interconversion between intestinal stem cell populations in distinct niches. Science 334, 1420–1424 (2011).
Powell, A. E. et al. The Pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158 (2012).
Muñoz, J. et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘ + 4’ cell markers. EMBO J. 31, 3079–3091 (2012).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Tye, H. et al. STAT3-driven upregulation of TLR2 promotes gastric tumorigenesis independent of tumor inflammation. Cancer Cell 22, 466–478 (2012).
Rakoff-Nahoum, S., Paglino, J., Eslami-Varzaneh, F., Edberg, S. & Medzhitov, R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118, 229–241 (2004).
Humphreys, R. C. et al. Apoptosis in the terminal endbud of the murine mammary gland: a mechanism of ductal morphogenesis. Development 122, 4013–4022 (1996).
Kitajima, S., Takuma, S. & Morimoto, M. Changes in colonic mucosal permeability in mouse colitis induced with dextran sulfate sodium. Exp. Anim. 48, 137–143 (1999).
Mailleux, A. et al. BIM regulates apoptosis during mammary ductal morphogenesis, and its absence reveals alternative cell death mechanisms. Dev. Cell 12, 221–234 (2007).
Hall, P. A., Coates, P. J., Ansari, B. & Hopwood, D. Regulation of cell number in the mammalian gastrointestinal tract: the importance of apoptosis. J. Cell Sci. 107, 3569–3577 (1994).
Bianchi, M. E. & Manfredi, A. A. High-mobility group box 1 (HMGB1) protein at the crossroads between innate and adaptive immunity. Immunol. Rev. 220, 35–46 (2007).
Park, J. S. et al. Involvement of toll-like receptors 2 and 4 in cellular activation by high mobility group box 1 protein. J. Biol. Chem. 279, 7370–7377 (2004).
Urbonaviciute, V. et al. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J. Exp. Med. 205, 3007–3018 (2008).
Schwitalla, S. et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38 (2012).
Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. Stem cells, cancer, and cancer stem cells. Nature 414, 105–111 (2001).
Barker, N. et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457, 608–611 (2009).
Shackleton, M., Quintana, E., Fearon, E. R. & Morrison, S. J. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 138, 822–829 (2009).
Ruark, E. et al. Mosaic PPM1D mutations are associated with predisposition to breast and ovarian cancer. Nature 493, 406–410 (2012).
Schepers, A. G. et al. Lineage tracing reveals Lgr5 + stem cell activity in mouse intestinal adenomas. Science 337, 730–735 (2012).
Dent, R. et al. Pattern of metastatic spread in triple-negative breast cancer. Breast Cancer Res. Treat. 115, 423–428 (2009).
Bauer, K. R., Brown, M., Cress, R. D., Parise, C. A. & Caggiano, V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: a population-based study from the California cancer Registry. Cancer 109, 1721–1728 (2007).
Carey, L. A. et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA 295, 2492–2502 (2006).
Willingham, S. B. et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl Acad. Sci. USA 109, 6662–6667 (2012).
Chao, M. P. et al. Calreticulin is the dominant pro-phagocytic signal on multiple human cancers and is counterbalanced by CD47. Sci. Transl. Med. 2, 63ra94 (2010).
Asselin-Labat, M-L. et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nat. Cell Biol. 9, 201–209 (2007).
el Marjou, F. et al. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39, 186–193 (2004).
Rothenberg, M. E. et al. Identification of a cKit(+) colonic crypt base secretory cell that supports Lgr5(+) stem cells in mice. Gastroenterology 142, 1195–1205.e6 (2012).
Dalerba, P. et al. Phenotypic characterization of human colorectal cancer stem cells. Proc. Natl Acad. Sci. USA 104, 10158–10163 (2007).
Shimono, Y. et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell 138, 592–603 (2009).
Hu, Y. & Smyth, G. K. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 347, 70–78 (2009).
Ran, F. A. et al. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 (2013).
Acknowledgements
This study was supported by the National Institutes of Health (NCI), the Breast Cancer Research Foundation, the Ludwig Institute, The California Institute for Regenerative Medicine and the Department of Defense (DOD). F.A.S. was supported by NWO-Rubicon grant, a fellowship from the Dutch Cancer Society and by a seed grant of the organization My Blue Dots. We thank T.N. Schumacher and M.A. Child for scientific input, S. Sim for her assistance with single-cell PCR assays, P. Lovelace for her assistance with flow cytometry and K. Montgomery for IHC. Some research was performed on a FACS Aria that was purchased using NIH S10 Shared Instrumentation Grant (1S10RR02933801) funds.
Author information
Authors and Affiliations
Contributions
F.A.S. and A.H.K. performed, designed and analysed research and wrote the paper; L.J.v.W., S.C., M.Z., S.S.S., I.G., D.J. and M.E.R. performed, designed and analysed research; D.Q. and J.S.L. performed research; J.P.V., T.K., S.R.Q., D.S. and M.v.d.R. designed and analysed research; F.M.D. and G.S. provided critical reagents; M.F.C. designed research and wrote paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 TLR2 and MyD88 expression in murine mammary epithelial subpopulations.
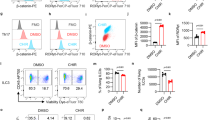
(a) Representative FACS dot plot and histogram of CD24, CD49f and CD61. Data are representative of 6 mice. (b) Quantitative rt-PCR on MEC populations as indicated in a. Basal markers: Krt5, Krt14, Krt17; Luminal marker: Krt19; N = 3 mice; ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.005; ∗∗∗∗P < 0.001. Error bars represent s.e.m. One-tail unpaired t-test analysis was used. (c) Ma-CFCs were sorted based on expression of CD24highCD49flow/neg luminal phenotype in combination with CD14. 200 cells luminal cells sorted for CD14pos or CD14neg were plated out on matrigel in triplicates. After 7-12 days colonies were counted and passaged as a single cell suspension again (N = 4 mice). 4 independent experiments were performed and the average is shown, P = 0.7, 0.9 (NS: non-significant). Values represent mean ± s.d. Student’s unpaired t-test for independent samples was used.
Supplementary Figure 2 Phenotypic characterization of mammary epithelial cells.
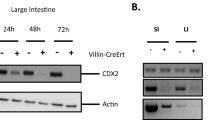
(a) Flow cytometric analysis of mammary epithelial cells from 6 weeks old mice of wild-type, Tlr2−/− and Cd14−/− mice. Cells were gated on life and lineage negative cells. (b) Immunohistochemistry on mammary glands from 6 weeks old mice of wild-type, Tlr2−/− and Cd14−/− mice. Myoepithelial cells (Cytokertain-14 (KRT14)) and luminal cells (E-CADHERIN) are present and properly organized in both wild-type as well as the knock out mammary glands. Scale bar is 50 μm. (c) Carmine Alum staining on whole mount of lactation of wild-type, Tlr2−/− and Cd14−/− mice. Scale bar is 1 mm. (d) Flow cytomety analysis of mammary epithelial cells from 6 to 8 weeks old mice. Cells were gated on live and lineage negative and stained for CD24, CD49f and CD14, TLR4, IL-1R1 or IL-18R1. In red is the isotype control for each specific sub-population, in blue is the CD14, TLR4, IL-1R1 or IL-18R1 staining. (e) Flow cytometry analysis of Myd88−/− mammary epithelial cells, from 6 weeks old mice of Krt14-CrenegMyd88f/f and Krt14-CreposMyd88f/f mice. Cells were gated on life and lineage negative cells. (f) Immunohistochemistry on mammary glands from 6 weeks old mice of Krt14-CrenegMyd88f/f and Krt14-CreposMyd88f/f mice. Myoepithelial cells (Cytokertain-14 (KRT14)) and luminal cells (E-CADHERIN) are present and properly organized in the MyD88 knockout mammary gland. Scale bar is 50 μm. All analyses were done with at least 3 mice.
Supplementary Figure 3 Limiting dilutions of mammary epithelial cells.
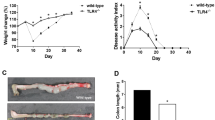
To determine the MRU frequency of each donor genotype we injected sorted cells, as indicated from donor mice of 10 to 14 weeks old mice in cleared mammary fat pads of three weeks old recipients. Recipient mice were syngeneic C57BL/6J mice. Numbers of successful out growths and numbers of total injections are shown for each dilution and genotype. MRU frequency and confidence was determined by ELDA graph and analysis. (a) Raw numbers for limiting dilutions of Tlr2−/− and Cd14−/− linneg MECs for Fig. 4a. (b,c) Raw numbers for limiting dilutions of Tlr4−/− linneg MECs, including ELDA analysis. Data for WT (N = 25 samples), Tlr4−/− (N = 30 samples) pooled from 3 independent experiments. P = 0.0085. (d) Raw numbers for limiting dilutions of Il-1r1−/− (N = 74 samples) and Il-18r1−/− linneg (N = 39 samples) MECs. (e,f) Limiting dilutions of Ink4a-Arf+/+Tlr2+/+(WT), Ink4a-Arf+/+Tlr2−/− (Tlr2KO), Ink4a-Arf−/−Tlr2+/+ (Ink4a-Arf KO) and Ink4a-Arf−/− Tlr2−/− (Ink4a-Arf KO Tlr2 KO) linneg MECs, including ELDA graph and analysis. Data for WT (N = 41 samples), Tlr2−/− KO (N = 75 samples), P < 0.0001, Ink4a-Arf KO (N = 66 samples), P = N.S. and Ink4a-Arf KO Tlr2 KO (N = 45 samples), P = N.S. are pooled from 4 independent experiments.
Supplementary Figure 4 Single cell gene expression of MRU.
Single cell expression showing expression of wild-type (black) and Tlr2−/− (red) MRUs for Fig. 3e. Cells were double sorted and subjected to multiplexed single cell rt-PCR. Normalized Ct values as visualized by comparing the distribution of Ct values in histograms. (a) Gene that was significantly up-regulated in Tlr2−/− cells as compared to wild-type cells. (b) Genes that were significantly down-regulated in Tlr2−/− as compared to wild-type cells. Each analysis was done on 2 different mice.
Supplementary Figure 5 Antibody and/or shRNA mediated blockade of TLR2, CD14, MYD88 and IRAK1.
(a) Representative examples colony formation on matrigel of primary ERneg breast cancer cells (N = 4). Scale bar is 20 μm. Cells were treated with control antibody or with neutralizing TLR2 and CD14 antibodies. (b,c) Two breast cell lines were transduced with the lentivirus to knockdown TLR2 and after puromycin selection cells were plated out and stained with crystal violet blue after 9 days in culture. Data are representative of 2 experiments. Scale bar is 1 cm. (d) MDA-MD-231 cells were transduced with control or indicated shRNA and knock-down efficiency was determined by qPCR. All shRNA mediated knock-down constructs resulted in a 4 to 12x reduction of its target mRNA. Data are representative of 2 experiments. (e) Knock-down of MYD88 and IRAK1 in the breast cell line MCF7. After transduction cells were sorted for Cherry and plated out on matrigel in triplicate to determine colony forming capacity. The number of colonies on matrigel is shown and a representative photo of the colonies is shown. N = 6 samples pooled from 2 independent experiments. ∗∗∗P < 0.001. Scale bar is 20 μm. Values represent mean ± s.d. Student’s unpaired t-test for independent samples was used. (f) MYD88 and IRAK1 knock down resulted in decreased expression of the NF-κB target gene IL-1B indicating that the knock down resulted in decreased NF-κB activity. Data are representative of 2 experiments.
Supplementary Figure 6 TLR2 immunohistochemistry on normal and tumour colon tissue.
(a) 10X magnification of immunohistochemistry for TLR2. Boxed is normal colon tissue, the rest are colon tumours. Scale bar is 200 μm. (b) 40X magnification of immunohistochemistry for TLR2 of colon tumours. Scale bar is 50 μm.
Supplementary Figure 7 Genome editing the Tlr2 locus using CAS9.
(a) Schematic of the targeted region of human TLR2 locus and targeting strategy. sgRNA cuts around + 944. The homologues recombination (HR) cassette has a left arm (including the E283* mutation) and a right arm. In between there is a CMV Cherry pA cassette (yellow box) in the anti sense orientation. Primer pairs (Pp1 & Pp2) are shown to detect homologues recombination. (b) Region of the sgRNA complementary to the protospacer. (c) Sequenced region show correct integration of HR construct as determined by sequencing the genomic junctions.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1156 kb)
Rights and permissions
About this article
Cite this article
Scheeren, F., Kuo, A., van Weele, L. et al. A cell-intrinsic role for TLR2–MYD88 in intestinal and breast epithelia and oncogenesis. Nat Cell Biol 16, 1238–1248 (2014). https://doi.org/10.1038/ncb3058
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb3058
This article is cited by
-
Commensal bacteria weaken the intestinal barrier by suppressing epithelial neuropilin-1 and Hedgehog signaling
Nature Metabolism (2023)
-
Characterization of a small molecule modulator of inflammatory cytokine production
Translational Medicine Communications (2022)
-
Context-dependent functions of pattern recognition receptors in cancer
Nature Reviews Cancer (2022)
-
Orchestration of myeloid-derived suppressor cells in the tumor microenvironment by ubiquitous cellular protein TCTP released by tumor cells
Nature Immunology (2021)
-
Synthetic lethality between MyD88 loss and mutations in Wnt/β-catenin pathway in intestinal tumor epithelial cells
Oncogene (2021)