Abstract
Pancreatic ductal adenocarcinoma (PDA) develops through distinct precursor lesions, including pancreatic intraepithelial neoplasia (PanIN) and intraductal papillary mucinous neoplasia (IPMN). However, genetic features resulting in IPMN-associated PDA (IPMN–PDA) versus PanIN-associated PDA (PanIN-PDA) are largely unknown. Here we find that loss of Brg1, a core subunit of SWI/SNF chromatin remodelling complexes, cooperates with oncogenic Kras to form cystic neoplastic lesions that resemble human IPMN and progress to PDA. Although Brg1-null IPMN–PDA develops rapidly, it possesses a distinct transcriptional profile compared with PanIN-PDA driven by mutant Kras and hemizygous p53 deletion. IPMN–PDA also is less lethal, mirroring prognostic trends in PDA patients. In addition, Brg1 deletion inhibits Kras-dependent PanIN development from adult acinar cells, but promotes Kras-driven preneoplastic transformation in adult duct cells. Therefore, this study implicates Brg1 as a determinant of context-dependent Kras-driven pancreatic tumorigenesis and suggests that chromatin remodelling may underlie the development of distinct PDA subsets.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Accession codes
References
Matthaei, H., Schulick, R. D., Hruban, R. H. & Maitra, A. Cystic precursors to invasive pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 8, 141–150 (2011).
Matthaei, H. et al. Clinicopathological characteristics and molecular analyses of multifocal intraductal papillary mucinous neoplasms of the pancreas. Ann. Surg. 255, 326–333 (2012).
Poultsides, G. A. et al. Histopathologic basis for the favorable survival after resection of intraductal papillary mucinous neoplasm-associated invasive adenocarcinoma of the pancreas. Ann. Surg. 251, 470–476 (2010).
Mino-Kenudson, M. et al. Prognosis of invasive intraductal papillary mucinous neoplasm depends on histological and precursor epithelial subtypes. Gut 60, 1712–1720 (2011).
Shi, C. & Hruban, R. H. Intraductal papillary mucinous neoplasm. Hum. Pathol. 43, 1–16 (2012).
Ray, K. C. et al. Epithelial tissues have varying degrees of susceptibility to Kras(G12D)-initiated tumorigenesis in a mouse model. PLoS One 6, e16786 (2011).
Visvader, J. E. Cells of origin in cancer. Nature 469, 314–322 (2011).
Habbe, N. et al. Spontaneous induction of murine pancreatic intraepithelial neoplasia (mPanIN) by acinar cell targeting of oncogenic Kras in adult mice. Proc Natl Acad. Sci. USA 105, 18913–18918 (2008).
Kopp, J. L. et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 22, 737–750 (2012).
Morris, J. P. t., Wang, S. C. & Hebrok, M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat. Rev. Cancer 10, 683–695 (2010).
von Figura, G., Morris, J. P. t., Wright, C. V. & Hebrok, M. Nr5a2 maintains acinar cell differentiation and constrains oncogenic Kras-mediated pancreatic neoplastic initiation. Gut (2013)10.1136/gutjnl-2012-304287
McKenna, E. S. & Roberts, C. W. Epigenetics and cancer without genomic instability. Cell Cycle 8, 23–26 (2009).
Wilson, B. G. & Roberts, C. W. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492 (2011).
Medina, P. P. & Sanchez-Cespedes, M. Involvement of the chromatin-remodelling factor BRG1/SMARCA4 in human cancer. Epigenetics 3, 64–68 (2008).
Versteege, I. et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394, 203–206 (1998).
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Li, M. et al. Inactivating mutations of the chromatin remodelling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 (2011).
Gui, Y. et al. Frequent mutations of chromatin remodelling genes in transitional cell carcinoma of the bladder. Nat. Genet. 43, 875–878 (2011).
Wiegand, K. C. et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 (2010).
Biankin, A. V. et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 491, 399–405 (2012).
Roberts, C. W., Leroux, M. M., Fleming, M. D. & Orkin, S. H. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumour suppressor gene Snf5. Cancer Cell 2, 415–425 (2002).
Roberts, C. W., Galusha, S. A., McMenamin, M. E., Fletcher, C. D. & Orkin, S. H. Haploinsufficiency of Snf5 (integrase interactor 1) predisposes to malignant rhabdoid tumours in mice. Proc Natl Acad. Sci. USA 97, 13796–13800 (2000).
Glaros, S., Cirrincione, G. M., Palanca, A., Metzger, D. & Reisman, D. Targeted knockout of BRG1 potentiates lung cancer development. Cancer Res. 68, 3689–3696 (2008).
Jones, S. et al. Core signalling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321, 1801–1806 (2008).
Shain, A. H. et al. Convergent structural alterations define SWItch/Sucrose NonFermentable (SWI/SNF) chromatin remodeler as a central tumour suppressive complex in pancreatic cancer. Proc Natl Acad. Sci. USA 109E, 252–259 (2012).
Dal Molin, M. et al. Loss of expression of the SWI/SNF chromatin remodelling subunit BRG1/SMARCA4 is frequently observed in intraductal papillary mucinous neoplasms of the pancreas. Hum. Pathol. 43, 585–591 (2012).
Sumi-Ichinose, C., Ichinose, H., Metzger, D. & Chambon, P. SNF2β-BRG1 is essential for the viability of F9 murine embryonal carcinoma cells. Mol. Cell Biol. 17, 5976–5986 (1997).
Kawaguchi, Y. et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32, 128–134 (2002).
Heiser, P. W., Lau, J., Taketo, M. M., Herrera, P. L. & Hebrok, M. Stabilization of β-catenin impacts pancreas growth. Development 133, 2023–2032 (2006).
Bardeesy, N. et al. Both p16(Ink4a) and the p19(Arf)-p53 pathway constrain progression of pancreatic adenocarcinoma in the mouse. Proc Natl Acad. Sci. USA 103, 5947–5952 (2006).
Hingorani, S. R. et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 4, 437–450 (2003).
Izeradjene, K. et al. Kras(G12D) and Smad4/Dpc4 haploinsufficiency cooperate to induce mucinous cystic neoplasms and invasive adenocarcinoma of the pancreas. Cancer Cell 11, 229–243 (2007).
Siveke, J. T. et al. Concomitant pancreatic activation of Kras(G12D) and Tgfa results in cystic papillary neoplasms reminiscent of human IPMN. Cancer Cell 12, 266–279 (2007).
Morris, J. P. t., Cano, D. A., Sekine, S., Wang, S. C. & Hebrok, M. β-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J. Clin. Invest. 120, 508–520 (2010).
Tanaka, T. et al. Evaluation of SOX9 expression in pancreatic ductal adenocarcinoma and intraductal papillary mucinous neoplasm. Pancreas 42, 488–493 (2012).
Fukuda, A. et al. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell 19, 441–455 (2011).
Lesina, M. et al. Stat3/Socs3 activation by IL-6 transsignalling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell 19, 456–469 (2011).
Corcoran, R. B. et al. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 71, 5020–5029 (2011).
Aguirre, A. J. et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 17, 3112–3126 (2003).
Morton, J. P. et al. LKB1 haploinsufficiency cooperates with Kras to promote pancreatic cancer through suppression of p21-dependent growth arrest. Gastroenterology 139, 586–597 (2010).
Takehara, A. et al. Gamma-aminobutyric acid (GABA) stimulates pancreatic cancer growth through overexpressing GABAA receptor pi subunit. Cancer Res. 67, 9704–9712 (2007).
Fusco, A. & Fedele, M. Roles of HMGA proteins in cancer. Nat. Rev. Cancer 7, 899–910 (2007).
Winslow, M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011).
Piscuoglio, S. et al. HMGA1 and HMGA2 protein expression correlates with advanced tumour grade and lymph node metastasis in pancreatic adenocarcinoma. Histopathology 60, 397–404 (2012).
Dozynkiewicz, M. A. et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev. Cell 22, 131–145 (2012).
Masui, T. et al. Expression of METH-1 and METH-2 in pancreatic cancer. Clin Cancer Res. 7, 3437–3443 (2001).
Wu, J. et al. Recurrent GNAS mutations define an unexpected pathway for pancreatic cyst development. Sci. Trans. Med. 3, 92ra66 (2011).
Wu, J. et al. Whole-exome sequencing of neoplastic cysts of the pancreas reveals recurrent mutations in components of ubiquitin-dependent pathways. Proc. Nat. Acad. Sci. USA 108, 21188–21193 (2011).
Furukawa, T. et al. Whole-exome sequencing uncovers frequent GNAS mutations in intraductal papillary mucinous neoplasms of the pancreas. Sci. Rep. 1, 161 (2011).
Kanda, M. et al. Mutant GNAS detected in duodenal collections of secretin-stimulated pancreatic juice indicates the presence or emergence of pancreatic cysts. Gut 62, 1024–1033 (2013).
Pan, F. C. et al. Spatiotemporal patterns of multipotentiality in Ptf1a-expressing cells during pancreas organogenesis and injury-induced facultative restoration. Development 140, 751–764 (2013).
Solar, M. et al. Pancreatic exocrine duct cells give rise to insulin-producing β cells during embryogenesis but not after birth. Dev. Cell 17, 849–860 (2009).
Wang, X. et al. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumours. Int. J. Cancer 112, 636–642 (2004).
Kang, H., Cui, K. & Zhao, K. BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol. Cell Biol. 24, 1188–1199 (2004).
Guan, B., Wang, T. L. & Shih Ie, M. ARID1A, a factor that promotes formation of SWI/SNF-mediated chromatin remodelling, is a tumour suppressor in gynecologic cancers. Cancer Res. 71, 6718–6727 (2011).
Hendricks, K. B., Shanahan, F. & Lees, E. Role for BRG1 in cell cycle control and tumour suppression. Mol. Cell Biol. 24, 362–376 (2004).
Kia, S. K., Gorski, M. M., Giannakopoulos, S. & Verrijzer, C. P. SWI/SNF mediates polycomb eviction and epigenetic reprogramming of the INK4b-ARF-INK4a locus. Mol. Cell Biol. 28, 3457–3464 (2008).
Bourgo, R. J. et al. SWI/SNF deficiency results in aberrant chromatin organization, mitotic failure, and diminished proliferative capacity. Mol. Biol. Cell 20, 3192–3199 (2009).
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Kent, W. J. et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank C. Wright for sharing Ptf1a–Cre and Ptf1a–CreER mice, D. Tuveson for KrasG12D mice, and P. Chambon and D. Reisman for Brg1flox mice, respectively. We thank C. Austin and D. Ngow for tissue processing and excellent technical assistance and all M.H. laboratory members for helpful discussion. Work in M.H.’s laboratory was supported by a grant from the NIH (CA112537). G.v.F. was supported by a post-doctoral Research Fellowship from the Deutsche Forschungsgemeinschaft (DFG, FI 1719/1-1) and a Klein Family Foundation Fellowship. A.F. was supported by a post-doctoral Research Fellowship from the Japan Society for the Promotion of Science, a Fellowship from the US National Pancreas Foundation, and a Fellowship from the Kato Memorial Biosciences Foundation. M.E.L. was supported by CIRM training grant TG2 01153. W.F.M., A.B. and K.J.H. were supported by a grant from the NIH (CA149548). Image acquisition was supported by the imaging core of the UCSF Diabetes and Endocrinology Research Center (DERC) NIH grant P30DK63720.
Author information
Authors and Affiliations
Contributions
G.V.F., A.F., N.R. and M.E.L. contributed to equal parts. G.V.F., A.F., N.R. and M.E.L. carried out all experiments and were involved together with M.H. in design and analysis of the experiments. G.V.F., A.F., N.R., M.E.L. and M.H. drafted the manuscript. J.P.M.I.V. generated cell lines, contributed to the survival analysis, was involved in experimental analysis, and critically reviewed the manuscript. G.E.K. performed the histopathological analysis including IPMN and tumour identification and tumour grading. H.R. performed quantification of tumour proliferation. J.F. generated the HNF1b–CreERT2 mice. D.W.D. analysed Brg1 expression on human samples. M.A.F., S.J.M. and J.F. provided intellectual contribution to this study. M.F.W., A.B. and K.J.H. carried out deep sequencing analyses. M.H. conceived the study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Cystic lesions in Ptf1a-Cre; KrasG12D; Brg1f/f pancreata are marked by thin stroma and are reminiscent of human pancreatobiliary IPMN.
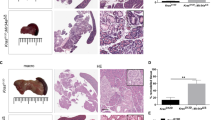
(a) Representative H&E staining of a cystic lesion in Ptf1a-Cre; KrasG12D; Brg1f/f pancreata reveals thin underlying stroma (S = stroma, E = epithelium). Despite some variability the majority of cystic lesions in Ptf1a-Cre; KrasG12D; Brg1f/f mice presented with thin stroma lacking cells with wavy nuclei. (b) H&E staining of representative fibrovascular bundle in Ptf1a-Cre; KrasG12D; Brg1f/f pancreata. (c) Cystic lesions of Ptf1a-Cre; KrasG12D; Brg1f/f and PanINs of Ptf1a-Cre; KrasG12D mice stain positive for Muc5AC (a’–e’), and Muc1 (a”’–e”’), but are negative for Muc2 (a”–e”). In contrast, ducts of control mice do not express Muc5AC and Muc2 (a’, a”, a”’). The mucin expression pattern of the cystic lesions in Ptf1a-Cre; KrasG12D; Brg1f/f pancreata (positivity for Muc1, and Muc5AC and negativity for Muc2) matches that of human IPMNs of the pancreatobiliary type (e’, e”, e”’). (a) and (c) scale bar 50 μm, (b) scale bar 100 μm.
Supplementary Figure 2 Brg1 is lost in neoplastic epithelium of Ptf1a-Cre; KrasG12D; Brg1f/f mice and characterization of Brg1 null PDA cell lines.
(a) (a’) Immunohistochemistry staining for Brg1 on neoplastic epithelium of a 9 weeks old Ptf1a-Cre; KrasG12D; Brg1f/f mouse. Low grade dysplasia marked by the presence of abundant mucin, undulating base, nuclear enlargement, or papillary or very dilated structures, tended to be negative for Brg1. In contrast, intermediate to high-grade dysplasia was uniformly negative for Brg1. (b’) Higher magnification of a low-grade dysplastic epithelium. (c’) Higher magnification of an intermediate to high-grade dysplastic epithelium. Scale bar 250 μm. (b) (a”) PCR analysis of the KrasG12D (Kras PCR: 1–4) and Brg1f/f (Brg1 PCR: 5–9) alleles in cancer cell lines. Murine genomic DNA was isolated from the following sources 1: Kras+/+ (embryonic fibroblasts isolated from a wild type mouse). 2: unrecombined KrasG12D/+ (embryonic fibroblasts isolated from a KrasG12D/+ mouse). 3: Ptf1a-Cre; KrasG12D; p53f/+ cancer cell line. 4: Ptf1a-Cre; KrasG12D; Brg1f/f cancer cell line. 5: Brg1+/+ (tail of a wild type mouse). 6: unrecombined Brg1f/+ (tail of Brg1f/+ mouse). 7: unrecombined Brg1f/f (tail of Brg1f/f mouse). 8: Ptf1a-Cre; KrasG12D; p53f/+ cancer cell line. 9: Ptf1a-Cre; KrasG12D; Brg1f/f cancer cell line. wt = wt allele, flox = unrecombined floxed allele, rec = recombined floxed allele. (b”) Western blot analysis of Brg1 in cancer cell lines derived from IPMN- and PanIN-PDAs. (c) Anoikis analysis of PanIN-PDA (n = 3), IPMN–PDA (n = 3 independent experiments) and Ptf1a-Cre; KrasG12D (n = 3 independent experiments) derived cancer cells by Annexin V/PI staining. 200,000 Cells were seeded onto poly-hema coated petri dishes to inhibit cell adhesion. After 48 h, detachment induced cell death or anoikis was assayed by measuring both early and late apoptosis. Total apoptosis is measured by counting both Annexin V single positive cells (early apoptotic) and Annexin V/PI double positive cells are (late apoptotic). Values are shown mean + /− SD. p value for total apoptosis was calculated by one way ANOVA between three sets of cell lines.
Supplementary Figure 3 Tumour suppressor gene expression in PDA and PDA precursor lesions.
(a) Immunohistochemistry staining for p53, p21, and p16 on pancreatic tissue isolated from PanIN-PDA and IPMN–PDA in mice. Scale bars 50 μm. (b) Immunohistochemistry for p53, p21, and p16 in ADM/PanIN and IPMN neoplastic precursor lesions on pancreatic sections derived from Ptf1a-Cre; KrasG12D and Ptf1a-Cre; KrasG12D; Brg1f/f mice. Insets show higher magnification pictures of PanIN or IPMN lesions. Scale bars 50 μm. (c) Quantification of p16 positive PDA cells in PanIN- versus IPMN–PDA (n = 7 tumours; values are shown as mean ± s.e.m. unpaired t-test was used for calculating p values). (d) Summary of tumour suppressor gene expression in cancer and precursor lesions of the respective genotypes. (e) Real-time PCR (RT-PCR) for Hmga2 relative to Cyclophilin A in murine pancreas containing PanIN (from Ptf1a-Cre; KrasG12D mice;n = 3) or IPMN (from Ptf1a-Cre; KrasG12D; Brg1f/f mice;n = 3) lesions. Values are shown mean ± s.e.m. Unpaired t-test was performed to calculate the p value.
Supplementary Figure 4 Brg1 null PDA cells display a gene pathway signature indicative of lower malignant potential.
Gene pathway/function analysis displaying the deep sequencing results of PanIN-PDA versus Brg1 null IPMN–PDA using Ingenuity®; software. The analysis was performed by focusing on those genes with significantly altered expression levels (p < 0.05) between PanIN- and IPMN–PDA. (a) Depicted is the heatmap clustering of the affected genes grouped into categories of cellular function. Highlighted in green are gene signatures with a z-score <= −2. The z-score reflects the significance and direction of the deviation of the individual gene signature from the mean. Category 1 = Cellular Movement, 2 = Hematological System Development and function, 3 = Cell to cell signalling and interaction, 4 = Tissue Development, 5 = Immune Cell Trafficking, 6 = Cancer, 7 = Cardiovascular system development and function, 8 = Inflammatory response, 9 = Cellular growth and proliferation, 10 = Cellular development, 11 = Organismal injury and abnormalities, 12 = Tissue morphology, 13 = Skeletal and muscular system development and function, 14 = Gastrointestinal diseases, 15 = Antigen presentation, 16 = Hepatic system disease, 17 = Infectious disease. (b) List of the 15 most significantly down-regulated pathways in IPMN–PDA.
Supplementary Figure 5 Sequence alignment of promoter regions.
Sequence alignment of promoter regions from mouse and human. Peak heights indicate degree of homology. Pink horizontal lines indicate evolutionary conserved regions. +1 indicates the start site. Black boxes are regions analyzed by ChIP. Blue: Coding exons, Yellow: Untranslated region, Red: Promoter elements, Salmon: Intronic region.
Supplementary Figure 6 ChIP analysis of promoter regions in PanIN- and IPMN–PDA cells.
(a) Relative fold enrichment of H3K4Me3 and H3K27Me3 (over IgG control) on promoter regions in PanIN-PDA cells (1 × 106 cells/ ., n = 3 independent experiments). Decreases in the solid color bars (H3K27) indicate a relative increase in active chromatin marks. Increases in the solid bars point to a relative increase in repressive marks. Each panel indicates individual cell lines. Values are shown as mean ± s.e.m. (b) Relative fold enrichment of H3K4Me3 and H3K27Me3 (over IgG control) on promoter regions in IPMN–PDA cells (1 × 106 cells/ ChIP; n = 3 independent experiments). Decreases in the solid color bars (H3K27) indicate a relative increase in active chromatin marks. Increases in the solid bars point to a relative increase in repressive marks. Each panel indicates individual cell lines. Values are shown as mean ± s.e.m.
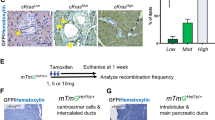
Supplementary Figure 7 Brg1 ablation abrogates PanIN formation from adult acinar cells and does not induce duct cell atypia in the absence of oncogenic Kras.
(a) H&E and Alcian blue stainings of pancreata derived from Ptf1a-CreER; Kras, Ptf1a-CreER; Kras; Brg1f/+ (= Brg1 het) and Ptf1a-CreER; Kras; Brg1f/f (= Brg1 KO) mice 4 months after tamoxifen induction. Note the strong reduction of Alcian blue PanIN lesions in Ptf1a-CreER; Kras; Brg1f/f (= Brg1 KO) mice. (b) A representative image of a pancreatic duct of an Hnf1b-CreERT2; Brg1f/f; R26REYFP mouse 6 weeks after tamoxifen induction. A total of, 3 Hnf1b-CreERT2; Brg1f/f (± R26REYFP) mice were analyzed 6 weeks (n = 1) or 12 weeks (n = 2) after tamoxifen induction. None of the mice showed duct cell atypia on histological examination (c) YFP staining confirmed recombination upon tamoxifen administration in both the large (arrow) and small (asterisks) duct system. (a) and (b) Scale bar 100 μm, (c) scale bar 50 μm.
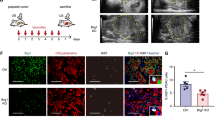
Supplementary Figure 8 Brg1 expression is associated with progression of human PanIN- and IPMN–PDA.
(a) Kaplan-Meier survival curve of PanIN-PDA patients with low or high Brg1 expression in tumour cells (n = 36 for low Brg1 and n = 106 for high Brg1). Brg1 expression was scored using a histoscore ranging from 0–8 (low to high expression). The cut off histoscore was 0–6 for low and 7–8 for high Brg1 expression. Log rank test, p = 0.007. Median survival was for low BRG1 = 15.1 months (95% CI 12.3-18.0) and for high BRG1 = 28.1 months (95% CI 24.3–31.8). (b) Brg1 labeling score from matched patient samples with IPMN and associated IPMN–PDA. The Brg1 expression was scored on the same section of a patient sample that contained an IPMN precursor and its associated IPMN–PDA. p value was calculated using the paired t-test; n=11 samples for IPMN precursors and n = 12 samples for IPMN–PDA, values are shown as mean ± s.e.m.
Supplementary Figure 9
Full scans.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1691 kb)
Supplementary Table 1
Supplementary Information (XLSX 13 kb)
Supplementary able 2
Supplementary Information (XLS 4130 kb)
Supplementary Table 3
Supplementary Information (XLS 35 kb)
Rights and permissions
About this article
Cite this article
von Figura, G., Fukuda, A., Roy, N. et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 16, 255–267 (2014). https://doi.org/10.1038/ncb2916
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2916
This article is cited by
-
Brg1 controls stemness and metastasis of pancreatic cancer through regulating hypoxia pathway
Oncogene (2023)
-
Smarcd3 is an epigenetic modulator of the metabolic landscape in pancreatic ductal adenocarcinoma
Nature Communications (2023)
-
Suppression of mutant Kirsten-RAS (KRASG12D)-driven pancreatic carcinogenesis by dual-specificity MAP kinase phosphatases 5 and 6
Oncogene (2022)
-
Molecular signaling in pancreatic ductal metaplasia: emerging biomarkers for detection and intervention of early pancreatic cancer
Cellular Oncology (2022)
-
Single-cell analysis of ductal differentiation
Nature Biomedical Engineering (2021)