Abstract
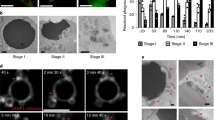
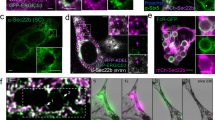
Autophagy normally involves the formation of double-membrane autophagosomes that mediate bulk cytoplasmic and organelle degradation. Here we report the modification of single-membrane vacuoles in cells by autophagy proteins. LC3 (Light chain 3) a component of autophagosomes, is recruited to single-membrane entotic vacuoles, macropinosomes and phagosomes harbouring apoptotic cells, in a manner dependent on the lipidation machinery including ATG5 and ATG7, and the class III phosphatidylinositol-3-kinase VPS34. These downstream components of the autophagy machinery, but not the upstream mammalian Tor (mTor)-regulated ULK–ATG13–FIP200 complex, facilitate lysosome fusion to single membranes and the degradation of internalized cargo. For entosis, a live-cell-engulfment program, the autophagy-protein-dependent fusion of lysosomes to vacuolar membranes leads to the death of internalized cells. As pathogen-containing phagosomes can be targeted in a similar manner, the death of epithelial cells by this mechanism mimics pathogen destruction. These data demonstrate that proteins of the autophagy pathway can target single-membrane vacuoles in cells in the absence of pathogenic organisms.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Kumari, S., Mg, S. & Mayor, S. Endocytosis unplugged: multiple ways to enter the cell. Cell Res. 20, 256–275 (2010).
Kawane, K. et al. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 292, 1546–1549 (2001).
Broadie, K. Axon pruning: an active role for glial cells. Curr. Biol. 14, R302–R304 (2004).
Elliott, M. R. & Ravichandran, K. S. Clearance of apoptotic cells: implications in health and disease. J. Cell Biol. 189, 1059–1070 (2010).
Savina, A. & Amigorena, S. Phagocytosis and antigen presentation in dendritic cells. Immunol. Rev. 219, 143–156 (2007).
Haas, A. The phagosome: compartment with a license to kill. Traffic 8, 311–330 (2007).
Steinberg, B. E. & Grinstein, S. Pathogen destruction versus intracellular survival: the role of lipids as phagosomal fate determinants. J. Clin. Invest. 118, 2002–2011 (2008).
Kinchen, J. M. et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat. Cell Biol. 10, 556–566 (2008).
Yang, Z. & Klionsky, D. J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 (2010).
Yang, Z. & Klionsky, D. J. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22, 124–131 (2010).
Jin, S. & White, E. Tumor suppression by autophagy through the management of metabolic stress. Autophagy 4, 563–566 (2008).
Burman, C. & Ktistakis, N. T. Autophagosome formation in mammalian cells. Semin. Immunopathol. 32, 397–413 (2010).
Levine, B. & Yuan, J. Autophagy in cell death: an innocent convict? J. Clin. Invest. 115, 2679–2688 (2005).
Rich, K. A., Burkett, C. & Webster, P. Cytoplasmic bacteria can be targets for autophagy. Cell Microbiol. 5, 455–468 (2003).
Yano, T. et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila. Nat. Immunol. 9, 908–916 (2008).
Gutierrez, M. G. et al. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 119, 753–766 (2004).
Nakagawa, I. et al. Autophagy defends cells against invading group A Streptococcus. Science 306, 1037–1040 (2004).
Ogawa, M. et al. Escape of intracellular Shigella from autophagy. Science 307, 727–731 (2005).
Ling, Y. M. et al. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J. Exp. Med. 203, 2063–2071 (2006).
Sanjuan, M. A., Milasta, S. & Green, D. R. Toll-like receptor signaling in the lysosomal pathways. Immunol. Rev. 227, 203–220 (2009).
Sanjuan, M. A. et al. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450, 1253–1257 (2007).
Overholtzer, M. et al. A nonapoptotic cell death process, entosis, that occurs by cell-in-cell invasion. Cell 131, 966–979 (2007).
Overholtzer, M. & Brugge, J. S. The cell biology of cell-in-cell structures. Nat. Rev. Mol. Cell Biol. 9, 796–809 (2008).
Seglen, P. O. & Gordon, P. B. 3-methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl Acad. Sci. USA 79, 1889–1892 (1982).
Ichimura, Y. et al. A ubiquitin-like system mediates protein lipidation. Nature 408, 488–492 (2000).
Hanada, T. et al. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 (2007).
Petiot, A., Ogier-Denis, E., Blommaart, E. F., Meijer, A. J. & Codogno, P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 275, 992–998 (2000).
Jung, C. H. et al. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 20, 1992–2003 (2009).
Hosokawa, N. et al. Nutrient-dependent mTORC1 association with theULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 20, 1981–1991 (2009).
Ganley, I. G. et al. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009).
Stenmark, H., Aasland, R., Toh, B. H. & D’Arrigo, A. Endosomal localization of the autoantigen EEA1 is mediated by a zinc-binding FYVE finger. J. Biol. Chem. 271, 24048–24054 (1996).
Eskelinen, E. L. et al. Inhibition of autophagy in mitotic animal cells. Traffic 3, 878–893 (2002).
Kimura, S., Noda, T. & Yoshimori, T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 (2007).
Sulston, J. E. & Horvitz, H. R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977).
Sulston, J. E., Schierenberg, E., White, J. G. & Thomson, J. N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 100, 64–119 (1983).
Ruck, A. et al. The Atg6/Vps30/Beclin1 ortholog BEC-1 mediates endocytic retrograde transport in addition to autophagy in C. elegans. Autophagy 7, 386–400 (2011).
Zhou, Z., Hartwieg, E. & Horvitz, H. R. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104, 43–56 (2001).
Kamada, Y., Sekito, T. & Ohsumi, Y. Autophagy in yeast: a TOR-mediated response to nutrient starvation. Curr. Top. Microbiol. Immunol. 279, 73–84 (2004).
Kim, D. H. et al. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 110, 163–175 (2002).
Mizushima, N. et al. A protein conjugation system essential for autophagy. Nature 395, 395–398 (1998).
Mizushima, N., Sugita, H., Yoshimori, T. & Ohsumi, Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 273, 33889–33892 (1998).
Hara, T. et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 (2008).
Cheong, H. et al. Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell 16, 3438–3453 (2005).
Kabeya, Y. et al. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16, 2544–2553 (2005).
Kamada, Y. et al. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513 (2000).
Zhao, Z. et al. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe 4, 458–469 (2008).
Lee, H. K. et al. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity 32, 227–239 (2010).
Qu, X. et al. Autophagy gene-dependent clearance of apoptotic cells during embryonic development. Cell 128, 931–946 (2007).
Huang, J. et al. Activation of antibacterial autophagy by NADPH oxidases. Proc. Natl Acad. Sci. USA 106, 6226–6231 (2009).
Shui, W. et al. Membrane proteomics of phagosomes suggests a connection to autophagy. Proc. Natl Acad. Sci. USA 105, 16952–16957 (2008).
Kabeya, Y. et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19, 5720–5728 (2000).
Weidberg, H. et al. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29, 1792–1802 (2010).
Fujita, N. et al. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell 19, 4651–4659 (2008).
Morvan, J. et al. In vitro reconstitution of fusion between immature autophagosomes and endosomes. Autophagy 5, 676–689 (2009).
Nakatogawa, H., Ichimura, Y. & Ohsumi, Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell 130, 165–178 (2007).
Weidberg, H. et al. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell 20, 444–454.
Nair, U. et al. SNARE proteins are required for macroautophagy. Cell 146, 290–302.
Behrends, C., Sowa, M. E., Gygi, S. P. & Harper, J. W. Network organization of the human autophagy system. Nature 466, 68–76 (2010).
Krajcovic, M. et al. A non-genetic route to aneuploidy in human cancers. Nat. Cell Biol. 13, 324–330 (2011).
Horbinski, C., Mojesky, C. & Kyprianou, N. Live free or die: tales of homeless (cells) in cancer. Am. J. Pathol. 177, 1044–1052 (2010).
Debnath, J. et al. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell 111, 29–40 (2002).
Debnath, J., Muthuswamy, S. K. & Brugge, J. S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 30, 256–268 (2003).
Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G. & Ahringer, J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, 1–10 (2000).
Acknowledgements
We thank J. Durgan, I. Ganley, X. Jiang, G. Mouneimne, E. Yao, A. Spencer and members of the Overholtzer laboratory for helpful discussions, reagents and for reading the manuscript. We also thank N. Lampen of the Memorial Sloan Kettering Cancer Center Electron Microscopy Facility for processing of electron microscopy samples. This work was financially supported by a grant from the National Cancer Institute (CA154649; M.O.), The Geoffrey Beene Cancer Research Center at MSKCC (M.O.), the Louis V. Gerstner, Jr. Young Investigators Fund (M.O. and C.M.H.) and the Alfred W. Bressler Scholar Fund (C.M.H.).
Author information
Authors and Affiliations
Contributions
O.F. and M.O. designed, carried out experiments and wrote the paper. S.E.K. and C.P.S. contributed experimental assistance and data. C.M.H. provided worm strains and carried out RNAi feeding of C. elegans.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1616 kb)
Supplementary Movie 1
Supplementary Information (AVI 905 kb)
Supplementary Movie 2
Supplementary Information (MOV 4035 kb)
Supplementary Movie 3
Supplementary Information (MOV 355 kb)
Supplementary Movie 4
Supplementary Information (AVI 14350 kb)
Supplementary Movie 5
Supplementary Information (MOV 6854 kb)
Supplementary Movie 6
Supplementary Information (AVI 1697 kb)
Supplementary Movie 7
Supplementary Information (AVI 1217 kb)
Supplementary Movie 8
Supplementary Information (AVI 1120 kb)
Supplementary Movie 9
Supplementary Information (MOV 896 kb)
Supplementary Movie 10
Supplementary Information (AVI 745 kb)
Supplementary Movie 11
Supplementary Information (MOV 1063 kb)
Rights and permissions
About this article
Cite this article
Florey, O., Kim, S., Sandoval, C. et al. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 13, 1335–1343 (2011). https://doi.org/10.1038/ncb2363
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2363
This article is cited by
-
Lysosomes as coordinators of cellular catabolism, metabolic signalling and organ physiology
Nature Reviews Molecular Cell Biology (2024)
-
Loss of RND3/RHOE controls entosis through LAMP1 expression in hepatocellular carcinoma
Cell Death & Disease (2024)
-
Knockdown of NEAT1 prevents post-stroke lipid droplet agglomeration in microglia by regulating autophagy
Cellular and Molecular Life Sciences (2024)
-
Autophagy and autophagy-related pathways in cancer
Nature Reviews Molecular Cell Biology (2023)
-
A guide to cell death pathways
Nature Reviews Molecular Cell Biology (2023)