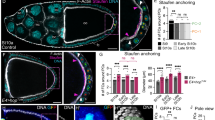
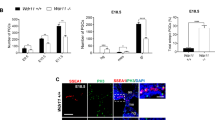
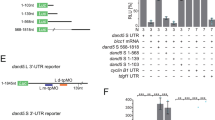
Abstract
Patterns of cell fates generated by morphogens are critically important for normal development; however, the mechanisms by which graded morphogen signals are converted into all-or-none cell fate responses are incompletely understood. In the Drosophila ovary, high and sustained levels of the secreted morphogen Unpaired (Upd) specify the migratory border-cell population by activating the signal transducer and activator of transcription1,2 (STAT). A lower or transient level of STAT activity specifies a non-migratory population of follicle cells3,4. Here we identify miR-279 as a component of a feedback pathway that further dampens the response in cells with low levels of JAK/STAT activity. miR-279 directly repressed STAT, and loss of miR-279 mimicked STAT gain-of-function or loss of Apontic (Apt), a known feedback inhibitor of STAT. Apt was essential for miR-279 expression in non-migratory follicle cells, whereas another STAT target, Ken and Barbie (Ken), downregulated miR-279 in border cells. Mathematical modelling and simulations of this regulatory circuit including miR-279, Apt and Ken supported key roles for miR-279 and Apt in generating threshold responses to the Upd gradient.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Silver, D. L. & Montell, D. J. Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell 107, 831–841 (2001).
Ghiglione, C. et al. The Drosophila cytokine receptor Domeless controls border cell migration and epithelial polarization during oogenesis. Development 129, 5437–5447 (2002).
Xi, R., McGregor, J. R. & Harrison, D. A. A gradient of JAK pathway activity patterns the anterior–posterior axis of the follicular epithelium. Dev. Cell 4, 167–177 (2003).
Starz-Gaiano, M., Melani, M., Wang, X., Meinhardt, H. & Montell, D. J. Feedback inhibition of Jak/STAT signaling by apontic is required to limit an invasive cell population. Dev. Cell 14, 726–738 (2008).
Driever, W. & Nusslein-Volhard, C. The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell 54, 95–104 (1988).
Gould, A., Morrison, A., Sproat, G., White, R. A. & Krumlauf, R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 11, 900–913 (1997).
Gould, A., Itasaki, N. & Krumlauf, R. Initiation of rhombomeric Hoxb4 expression requires induction by somites and a retinoid pathway. Neuron 21, 39–51 (1998).
Saka, Y. & Smith, J. C. A mechanism for the sharp transition of morphogen gradient interpretation in Xenopus. BMC Dev. Biol. 7, 47 (2007).
Ghiglione, C., Devergne, O., Cerezo, D. & Noselli, S. Drosophila RalA is essential for the maintenance of Jak/Stat signalling in ovarian follicles. EMBO Rep. 9, 676–682 (2008).
Silver, D. L., Geisbrecht, E. R. & Montell, D. J. Requirement for JAK/STAT signaling throughout border cell migration in Drosophila. Development 132, 3483–3492 (2005).
Bartel, D. P. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 (2009).
Hornstein, E. & Shomron, N. Canalization of development by microRNAs. Nat. Genet. 38 (Suppl), S20–S24 (2006).
John, B. et al. Human microRNA targets. PLoS Biol. 2, e363 (2004).
Krek, A. et al. Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 (2005).
Lewis, B. P., Burge, C. B. & Bartel, D. P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 (2005).
Enright, A. J. et al. MicroRNA targets in Drosophila. Genome Biol. 5, R1 (2003).
Cayirlioglu, P. et al. Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319, 1256–1260 (2008).
Ebert, M. S., Neilson, J. R. & Sharp, P. A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 4, 721–726 (2007).
Loya, C. M., Lu, C. S., Van Vactor, D. & Fulga, T. A. Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat. Methods 6, 897–903 (2009).
Bach, E. A. et al. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr. Patterns 7, 323–331 (2007).
Starz-Gaiano, M., Melani, M., Meinhardt, H. & Montell, D. Interpretation of the UPD/JAK/STAT morphogen gradient in Drosophila follicle cells. Cell Cycle 8, 2917–2925 (2009).
Arbouzova, N. I., Bach, E. A. & Zeidler, M. P. Ken & barbie selectively regulates the expression of a subset of Jak/STAT pathway target genes. Curr. Biol. 16, 80–88 (2006).
Wang, X. et al. Analysis of cell migration using whole-genome expression profiling of migratory cells in the Drosophila ovary. Dev. Cell 10, 483–495 (2006).
Lukacsovich, T., Asztalos, Z., Juni, N., Awano, W. & Yamamoto, D. The Drosophila melanogaster 60A chromosomal division is extremely dense with functional genes: their sequences, genomic organization, and expression. Genomics 57, 43–56 (1999).
Castrillon, D. H. et al. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics 135, 489–505 (1993).
Flaherty, M. S., Zavadil, J., Ekas, L. A. & Bach, E. A. Genome-wide expression profiling in the Drosophila eye reveals unexpected repression of notch signaling by the JAK/STAT pathway. Dev. Dyn. 238, 2235–2253 (2009).
Schier, A. F. Nodal morphogens. Cold Spring Harb. Perspect. Biol. 1, a003459 (2009).
Baksa, K., Parke, T., Dobens, L. L. & Dearolf, C. R. The Drosophila STAT protein, stat92E, regulates follicle cell differentiation during oogenesis. Dev. Biol. 243, 166–175 (2002).
Singh, S. R., Liu, W. & Hou, S. X. The adult Drosophila malpighian tubules are maintained by multipotent stem cells. Cell Stem. Cell 1, 191–203 (2007).
Gellon, G., Harding, K. W., McGinnis, N., Martin, M. M. & McGinnis, W. A genetic screen for modifiers of Deformed homeotic function identifies novel genes required for head development. Development 124, 3321–3331 (1997).
Lee, T. & Luo, L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999).
Rorth, P. et al. Systematic gain-of-function genetics in Drosophila. Development 125, 1049–1057 (1998).
Manseau, L. et al. GAL4 enhancer traps expressed in the embryo, larval brain, imaginal discs, and ovary of Drosophila. Dev. Dyn. 209, 310–322 (1997).
Fischer, J. A., Giniger, E., Maniatis, T. & Ptashne, M. GAL4 activates transcription in Drosophila. Nature 332, 853–856 (1988).
Eulenberg, K. G. & Schuh, R. The tracheae defective gene encodes a bZIP protein that controls tracheal cell movement during Drosophila embryogenesis. EMBO J. 16, 7156–7165 (1997).
Pek, J. W., Lim, A. K. & Kai, T. Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7. Dev. Cell 17, 417–424 (2009).
Acknowledgements
This work was supported by NIH grant GM46425 to D.J.M. W.H.Y. was supported by a fellowship from the Korea Science and Engineering Foundation (KOSEF) and the H.A. and Mary K. Chapman Young Investigator Fellowship. We acknowledge A. H. McDonald and B. Steiner for technical assistance. We thank J. S. Kang for data illustration. We thank M. Issigonis for helpful discussion. We thank present and past members of the D.J.M. and C. Montell laboratories for helpful discussion and comments. Flybase and the Bloomington Drosophila Stock Center provided critical information and reagents for this study.
Author information
Authors and Affiliations
Contributions
W.H.Y. planned the experimental design, conducted the experiments and analysed data. H.M. developed and tested the mathematical model. D.J.M. conceived of the project, participated in experimental design, discussions of results and interpretations, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 829 kb)
Supplementary Movie 1
Supplementary Information (MOV 1613 kb)
Supplementary Movie 2
Supplementary Information (MOV 5345 kb)
Rights and permissions
About this article
Cite this article
Yoon, W., Meinhardt, H. & Montell, D. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nat Cell Biol 13, 1062–1069 (2011). https://doi.org/10.1038/ncb2316
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2316
This article is cited by
-
Type I Interferons in the Pathogenesis and Treatment of Autoimmune Diseases
Clinical Reviews in Allergy & Immunology (2020)
-
Identification of miRNAs and their target genes in Larix olgensis and verified of differential expression miRNAs
BMC Plant Biology (2019)
-
Cell motility in cancer invasion and metastasis: insights from simple model organisms
Nature Reviews Cancer (2018)
-
Apontic regulates somatic stem cell numbers in Drosophila testes
BMC Developmental Biology (2016)
-
A transgenic resource for conditional competitive inhibition of conserved Drosophila microRNAs
Nature Communications (2015)