Abstract
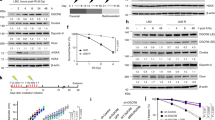
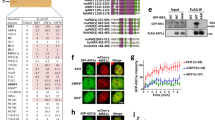
Regulatory ubiquitylation is emerging as an important mechanism to protect genome integrity in cells exposed to DNA damage1,2,3,4,5,6,7,8,9. However, the spectrum of known ubiquitin regulators of the DNA damage response (DDR) is limited and their functional interplay is poorly understood. Here, we identify HERC2 as a factor that regulates ubiquitin-dependent retention of repair proteins on damaged chromosomes. In response to ionising radiation (IR), HERC2 forms a complex with RNF8, a ubiquitin ligase involved in the DDR3,4,5,6. The HERC2–RNF8 interaction requires IR-inducible phosphorylation of HERC2 at Thr 4827, which in turn binds to the forkhead-associated (FHA) domain of RNF8. Mechanistically, we provide evidence that HERC2 facilitates assembly of the ubiquitin-conjugating enzyme Ubc13 with RNF8, thereby promoting DNA damage-induced formation of Lys 63-linked ubiquitin chains. We also show that HERC2 interacts with, and maintains the levels of, RNF168, another ubiquitin ligase operating downstream of RNF8 (Refs 7, 8). Consequently, knockdown of HERC2 abrogates ubiquitin-dependent retention of repair factors such as 53BP1, RAP80 and BRCA1. Together with the increased radiosensitivity of HERC2-depleted cells, these results uncover a regulatory layer in the orchestration of protein interactions on damaged chromosomes and they underscore the role of ubiquitin-mediated signalling in genome maintenance.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Change history
10 March 2010
In the version of this letter initially published online, an author Claudia Lukas, was omitted. This error has been corrected in both the HTML and PDF versions of the letter.
References
Bartek, J. & Lukas, J. DNA damage checkpoints: from initiation to recovery or adaptation. Curr. Opin. Cell Biol. 19, 238–245 (2007).
Bennett, E. J. & Harper, J. W. DNA damage: ubiquitin marks the spot. Nature Struct. Mol. Biol. 15, 20–22 (2008).
Huen, M. S. et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 (2007).
Kolas, N. K. et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 (2007).
Mailand, N. et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131, 887–900 (2007).
Wang, B. & Elledge, S. J. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl Acad. Sci. USA 104, 20759–20763 (2007).
Doil, C. et al. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136, 435–446 (2009).
Stewart, G. S. et al. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136, 420–434 (2009).
Jackson, S. P. & Bartek, J. The DNA damage response in human biology and disease. Nature 461, 1071–1078 (2009).
Ji, Y. et al. The ancestral gene for transcribed, low-copy repeats in the Prader-Willi/Angelman region encodes a large protein implicated in protein trafficking, which is deficient in mice with neuromuscular and spermiogenic abnormalities. Hum. Mol. Genet. 8, 533–542 (1999).
Lehman, A. L. et al. A very large protein with diverse functional motifs is deficient in rjs (runty, jerky, sterile) mice. Proc. Natl Acad. Sci. USA 95, 9436–9441 (1998).
Nicholls, R. D. & Knepper, J. L. Genome organization, function, and imprinting in Prader-Willi and Angelman syndromes. Annu. Rev. Genomics Hum. Genet. 2, 153–175 (2001).
Durocher, D., Henckel, J., Fersht, A. R. & Jackson, S. P. The FHA domain is a modular phosphopeptide recognition motif. Mol. Cell 4, 387–394 (1999).
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007).
Plans, V. et al. The RING finger protein RNF8 recruits UBC13 for lysine 63-based self polyubiquitylation. J. Cell Biochem. 97, 572–582 (2006).
Zhao, G. Y. et al. A critical role for the ubiquitin-conjugating enzyme Ubc13 in initiating homologous recombination. Mol. Cell 25, 663–675 (2007).
Ito, K. et al. N-terminally extended human ubiquitin-conjugating enzymes (E2s) mediate the ubiquitylation of RING-finger proteins, ARA54 and RNF8. Eur. J. Biochem. 268, 2725–2732 (2001).
Huen, M. S. et al. Noncanonical E2 variant-independent function of UBC13 in promoting checkpoint protein assembly. Mol. Cell Biol. 28, 6104–6112 (2008).
Rendtlew Danielsen, J. R. et al. HCLK2 is required for activity of the DNA damage response kinase ATR. J. Biol. Chem. 284, 4140–4147 (2009).
Takai, H. et al. Tel2 regulates the stability of PI3K-related protein kinases. Cell 131, 1248–1259 (2007).
Shevchenko, A., Wilm, M., Vorm, O. & Mann, M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 (1996).
Bekker-Jensen, S. et al. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173, 195–206 (2006).
Mailand, N., Bekker-Jensen, S., Bartek, J. & Lukas, J. Destruction of Claspin by SCFβTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 23, 307–318 (2006).
Acknowledgements
We thank Z. Ronai and D. Durocher for reagents, and J Chen for sharing unpublished data. This work was supported by grants from the Danish Cancer Society, Danish National Research Foundation, European Commission (integrated projects GENICA and DNA Repair), Lundbeck Foundation, Danish Research Council and the John and Birthe Meyer Foundation.
Author information
Authors and Affiliations
Contributions
N.M., S.B.-J., J.B. and J.L. designed the project and wrote the paper. N.M., S.B.-J. and J.R.D. performed most of the biochemical, cell biology and phosphorylation/clonogenic survival experiments, respectively. C.L. provided the in situ RNF168 data, I.G. performed the mass spectrometry analysis, A.N. generated the HERC2 constructs and K.F. contributed to the analysis of HERC2 complexes. All authors participated in interpreting the results.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information Figures (PDF 1220 kb)
Rights and permissions
About this article
Cite this article
Bekker-Jensen, S., Danielsen, J., Fugger, K. et al. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat Cell Biol 12, 80–86 (2010). https://doi.org/10.1038/ncb2008
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2008
This article is cited by
-
Mechanisms of Ferritinophagy and Ferroptosis in Diseases
Molecular Neurobiology (2023)
-
SRSF1 is essential for primary follicle development by regulating granulosa cell survival via mRNA alternative splicing
Cellular and Molecular Life Sciences (2023)
-
Genetic alterations associated with malignant transformation of sporadic vestibular schwannoma
Acta Neurochirurgica (2022)
-
HERC2 inactivation abrogates nucleolar localization of RecQ helicases BLM and WRN
Scientific Reports (2021)
-
RNA-seq analysis of galaninergic neurons from ventrolateral preoptic nucleus identifies expression changes between sleep and wake
BMC Genomics (2020)