Abstract
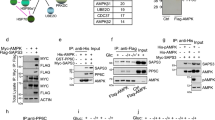
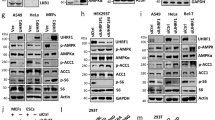
The fasting-activated longevity protein sirtuin 1 (SirT1, ref. 1) promotes gluconeogenesis in part, by increasing transcription of the key gluconeogenic genes pepck1 and g6pase2,3, through deacetylating PGC-1α and FOXO1 (ref. 4). In contrast, signal transducer and activator of transcription 3 (STAT3) inhibits glucose production by suppressing expression of these genes5,6. It is not known whether the inhibition of gluconeogenesis by STAT3 is controlled by metabolic regulation. Here we show that STAT3 phosphorylation and function in the liver were tightly regulated by the nutritional status of an animal, through SirT1-mediated deacetylation of key STAT3 lysine sites. The importance of the SirT1–STAT3 pathway in the regulation of gluconeogenesis was verified in STAT3-deficient mice in which the dynamic regulation of gluconeogenic genes by nutritional status was disrupted. Our results reveal a new nutrient sensing pathway through which SirT1 suppresses the inhibitory effect of STAT3, while activating the stimulatory effect of PGC-1α and FOXO1 on gluconeogenesis, thus ensuring maximal activation of gluconeogenic gene transcription. The connection between acetylation and phosphorylation of STAT3 implies that STAT3 may have an important role in other cellular processes that involve SirT1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Luo, J. et al. Negative control of p53 by Sir2α promotes cell survival under stress. Cell 107, 137–148 (2001).
Rodgers, J. T. et al. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434, 113–118 (2005).
Banks, A. S. et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 8, 333–341 (2008).
Frescas, D., Valenti, L. & Accili, D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 280, 20589–20595 (2005).
Inoue, H. et al. Role of STAT-3 in regulation of hepatic gluconeogenic genes and carbohydrate metabolism in vivo. Nature Med. 10, 168–174 (2004).
Inoue, H. et al. Role of hepatic STAT3 in brain-insulin action on hepatic glucose production. Cell Metab. 3, 267–275 (2006).
Zhong, Z., Wen, Z. & Darnell, J. E. Jr Stat3: a STAT family member activated by tyrosine phosphorylation in response to epidermal growth factor and interleukin-6. Science 264, 95–98 (1994).
Ray, S., Boldogh, I. & Brasier, A. R. STAT3 NH2-terminal acetylation is activated by the hepatic acute-phase response and required for IL-6 induction of angiotensinogen. Gastroenterology 129, 1616–1632 (2005).
Wang, R., Cherukuri, P. & Luo, J. Activation of Stat3 sequence-specific DNA binding and transcription by p300/CREB-binding protein-mediated acetylation. J. Biol. Chem. 280, 11528–11534 (2005).
Yuan, Z. L., Guan, Y. J., Chatterjee, D. & Chin, Y. E. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science 307, 269–273 (2005).
Rodgers, J. T., Lerin, C., Gerhart-Hines, Z. & Puigserver, P. Metabolic adaptations through the PGC-1α and SIRT1 pathways. FEBS Lett. 582, 46–53 (2008).
Chen, D. et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 22, 1753–1757 (2008).
Solomon, J. M. et al. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell. Biol. 26, 28–38 (2006).
Chan, J. H., Lim, S. & Wong, W. S. Antisense oligonucleotides: from design to therapeutic application. Clin. Exp. Pharmacol. Physiol. 33, 533–540 (2006).
Savage, D. B. et al. Reversal of diet-induced hepatic steatosis and hepatic insulin resistance by antisense oligonucleotide inhibitors of acetyl-CoA carboxylases 1 and 2. J. Clin. Invest. 116, 817–824 (2006).
Puigserver, P. et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423, 550–555 (2003).
Nakae, J., Park, B. C. & Accili, D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J. Biol. Chem. 274, 15982–15985 (1999).
Howitz, K. T. et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425, 191–196 (2003).
Li, X. et al. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol. Cell 28, 91–106 (2007).
Ray, S., Lee, C., Hou, T., Boldogh, I. & Brasier, A. R. Requirement of histone deacetylase1 (HDAC1) in signal transducer and activator of transcription 3 (STAT3) nucleocytoplasmic distribution. Nucleic Acids Res. (2008).
Becker, S., Groner, B. & Muller, C. W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature 394, 145–151 (1998).
Koch, C. A., Anderson, D., Moran, M. F., Ellis, C. & Pawson, T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science 252, 668–674 (1991).
Pawson, T. & Gish, G. D. SH2 and SH3 domains: from structure to function. Cell 71, 359–362 (1992).
Minami, M. et al. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc. Natl Acad. Sci. USA 93, 3963–3966 (1996).
Maloney, A. et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 67, 3239–3253 (2007).
Yoon, J. C. et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413, 131–138 (2001).
Acknowledgements
We thank M. Shanabrough for her technical support and careful revision of this manuscript. Some constructs were obtained from Y. E. Chin, W. Gu, P. Yao, E. Seto and D. Levy. SirT1, PGC-1α adenovirus was a gift from P. P. Puigserver. SirT1 KO MEFs and wild-type MEFs were a gift from L. P. Guarente. Part of this work was supported by an ADA grant to Q. G. (1-08-RA-54) and NIH grants to T. L. H. (DK-08000 and DK-060711), G. I. S. (DK-40936 and DK-076169) and J.L.B. (DK-P30-34989). The preparation of primary hepatocytes was performed in the Liver Center of Yale University School of Medicine. SirT1 ASO and Control ASO were provided by ISIS Pharmaceuticals, Inc.
Author information
Authors and Affiliations
Contributions
Y.N., T.L.H. and Q.G. designed, executed and analysed most of the experiments and wrote the paper. D.M.E. contributed to the execution of the SirT1-ASO animal experiments and edited the paper. Z.Y. contributed to construction of trunicated STAT3 plasmids. M.D. designed, performed and analysed the animal experiements with EX527 treatement. G.I.S. provided critical models and analysed the data of the animal experiments.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 2100 kb)
Rights and permissions
About this article
Cite this article
Nie, Y., Erion, D., Yuan, Z. et al. STAT3 inhibition of gluconeogenesis is downregulated by SirT1. Nat Cell Biol 11, 492–500 (2009). https://doi.org/10.1038/ncb1857
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1857
This article is cited by
-
Association between miR-138-5p, miR-132-3p, SIRT1, STAT3, and CD36 and atherogenic indices in blood mononuclear cells from patients with atherosclerosis
Egyptian Journal of Medical Human Genetics (2023)
-
Inherited myogenic abilities in muscle precursor cells defined by the mitochondrial complex I-encoding protein
Cell Death & Disease (2023)
-
Novel Therapeutic Avenues for Hypertrophic Cardiomyopathy
American Journal of Cardiovascular Drugs (2023)
-
Roles of sirtuin family members in chronic obstructive pulmonary disease
Respiratory Research (2022)
-
A novel resveratrol analog upregulates sirtuin 1 and inhibits inflammatory cell infiltration in acute pancreatitis
Acta Pharmacologica Sinica (2022)