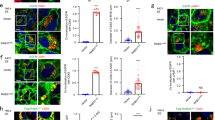
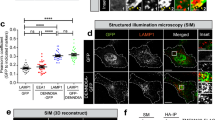
Abstract
Removal of apoptotic cells is critical for the physiological well-being of the organism1 and defects in corpse removal have been linked to disease states2,3. Genes regulating corpse recognition and internalization have been identified4, but few molecules involved in the processing of internalized corpses are known. Through a combination of targeted and unbiased reverse genetic screens in Caenorhabditis elegans, and studies in mammalian cells, we have identified genes required for maturation of apoptotic-cell-containing phagosomes. We have further ordered these candidates, which include the GTPases RAB-5 and RAB-7 and the HOPS complex, into a coherent linear pathway for the maturation of apoptotic cells within phagosomes. In depth analysis of two additional candidate genes, the phosphatidylinositol 3 kinase (PI(3)K) vps-34 (A001762) and dyn-1/dynamin, showed an accumulation of internalized, but undegraded, corpses within abnormal Rab5-negative phagosomes. We ordered these candidates in our pathway, with DYN-1 functioning upstream of VPS-34 in the recruitment and/or retention of RAB-5 to the phagosome. Finally, we have also identified a previously undescribed biochemical complex containing Vps34, dynamin and Rab5GDP, thus providing a mechanism for Rab5 recruitment to the nascent phagosome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ravichandran, K. S. & Lorenz, U. Engulfment of apoptotic cells: signals for a good meal. Nature Rev. Immunol. 7, 964–974 (2007).
Kawane, K. et al. Chronic polyarthritis caused by mammalian DNA that escapes from degradation in macrophages. Nature 443, 998–1002 (2006).
Schrijvers, D. M., De Meyer, G. R., Kockx, M. M., Herman, A. G. & Martinet, W. Phagocytosis of apoptotic cells by macrophages is impaired in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 25, 1256–1261 (2005).
Kinchen, J. M. & Hengartner, M. O. Tales of cannibalism, suicide, and murder: programmed cell death in C. elegans. Curr. Top. Dev. Biol. 65, 1–45 (2005).
Henson, P. M. & Hume, D. A. Apoptotic cell removal in development and tissue homeostasis. Trends Immunol. 27, 244–250 (2006).
Franz, S. et al. Apoptosis and autoimmunity: when apoptotic cells break their silence. Curr. Rheumatol. Rep. 8, 245–247 (2006).
Gaipl, U. S. et al. Inefficient clearance of dying cells and autoreactivity. Curr. Top. Microbiol. Immunol. 305, 161–176 (2006).
Lettre, G. & Hengartner, M. O. Developmental apoptosis in C. elegans: a complex CEDnario. Nature Rev. Mol. Cell Biol. 7, 97–108 (2006).
Gumienny, T. L., Lambie, E., Hartwieg, E., Horvitz, H. R. & Hengartner, M. O. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development 126, 1011–1022 (1999).
Kinchen, J. M. & Ravichandran, K. S. Journey to the grave: signaling events regulating removal of apoptotic cells. J. Cell Sci. 120, 2143–2149 (2007).
Mangahas, P. M. & Zhou, Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin. Cell Dev. Biol. 16, 295–306 (2005).
Gruenberg, J. & van der Goot, F. G. Mechanisms of pathogen entry through the endosomal compartments. Nature Rev. Mol. Cell Biol. 7, 495–504 (2006).
Grosshans, B. L., Ortiz, D. & Novick, P. Rabs and their effectors: achieving specificity in membrane traffic. Proc. Natl Acad. Sci. USA 103, 11821–11827 (2006).
Horazdovsky, B. F. & Emr, S. D. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J. Biol. Chem. 268, 4953–4962 (1993).
Sato, T. K., Rehling, P., Peterson, M. R. & Emr, S. D. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6, 661–671 (2000).
Vieira, O. V. et al. Distinct roles of class I and class III phosphatidylinositol 3-kinases in phagosome formation and maturation. J. Cell Biol. 155, 19–25 (2001).
Gengyo-Ando, K. et al. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 8, 152–157 (2007).
Hayakawa, A. et al. The WD40 and FYVE domain containing protein 2 defines a class of early endosomes necessary for endocytosis. Proc. Natl Acad. Sci. USA 103, 11928–11933 (2006).
Carney, D. S., Davies, B. A. & Horazdovsky, B. F. Vps9 domain-containing proteins: activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 16, 27–35 (2006).
Sato, M. et al. Caenorhabditis elegans RME-6 is a novel regulator of RAB-5 at the clathrin-coated pit. Nature Cell Biol. 7, 559–569 (2005).
Lettre, G. et al. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ. 11, 1198–1203 (2004).
Kinchen, J. M. et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature 434, 93–99 (2005).
Kamath, R. S. et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421, 231–237 (2003).
Thompson, H. M., Skop, A. R., Euteneuer, U., Meyer, B. J. & McNiven, M. A. The large GTPase dynamin associates with the spindle midzone and is required for cytokinesis. Curr. Biol. 12, 2111–2117 (2002).
Clark, S. G., Shurland, D. L., Meyerowitz, E. M., Bargmann, C. I. & van der Bliek, A. M. A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C. elegans. Proc. Natl Acad. Sci. USA 94, 10438–10443 (1997).
Yu, X., Odera, S., Chuang, C. H., Lu, N. & Zhou, Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev. Cell 10, 743–757 (2006).
Orth, J. D. & McNiven, M. A. Dynamin at the actin-membrane interface. Curr. Opin. Cell Biol. 15, 31–39 (2003).
Krueger, E. W., Orth, J. D., Cao, H. & McNiven, M. A. A dynamin-cortactin-Arp2/3 complex mediates actin reorganization in growth factor-stimulated cells. Mol. Biol. Cell 14, 1085–1096 (2003).
Schafer, D. A. Coupling actin dynamics and membrane dynamics during endocytosis. Curr. Opin. Cell Biol. 14, 76–81 (2002).
Gold, E. S. et al. Dynamin 2 is required for phagocytosis in macrophages. J. Exp. Med. 190, 1849–1856 (1999).
Christoforidis, S. et al. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nature Cell Biol. 1, 249–252 (1999).
Roggo, L. et al. Membrane transport in Caenorhabditis elegans: an essential role for VPS34 at the nuclear membrane. EMBO J. 21, 1673–1683 (2002).
Chang, H. C., Hull, M. & Mellman, I. The J-domain protein Rme-8 interacts with Hsc70 to control clathrin-dependent endocytosis in Drosophila. J. Cell Biol. 164, 1055–64 (2004).
Bieri, T. et al. WormBase: new content and better access. Nucleic Acids Res. 35, D506–D510 (2007).
Grimsley, C. M., Lu, M., Haney, L. B., Kinchen, J. M. & Ravichandran, K. S. Characterization of a novel interaction between ELMO1 and ERM proteins. J. Biol. Chem. 281, 5928–5937 (2006).
Tosello-Trampont, A. C. et al. Identification of two signaling submodules within the CrkII/ELMO/Dock180 pathway regulating engulfment of apoptotic cells. Cell Death Differ. 14, 963–972 (2007).
Lu, M. et al. PH domain of ELMO functions in trans to regulate Rac activation via Dock180. Nature Struct. Mol. Biol. 11, 756–762 (2004).
Acknowledgements
The authors would like to thank members of the Ravichandran and Hengartner laboratories for helpful input and suggestions. We thank Dorothy Schafer for dynamin plasmids and purified Dyn2 protein, David Castle, Jim Casanova and Heidi McBride for Rab5 expression constructs, Jae Jung for Vps34 expression constructs, Lukas Neukomm for pLN022 and pLN019 plasmids and Jan Redick and Christie Davis from the Advanced Microscopy Facility for help with confocal microscopy. Some strains used in this work were obtained from the Caenorhabditis Genetics Center (CGC). This work was supported by grants from the NIGMS/NIH to K. S. R. and from the EU Project Apoclear, the Ernst Hadorn Foundation, the University of Zurich and the Swiss National Science Foundation to M. O. H. J. M. K. is an Arthritis Foundation Postdoctoral Fellow.
Author information
Authors and Affiliations
Contributions
J. M. K., K. D., J. A. and L. S. contributed to the generation of nematode transgenics and fluorescence microscopy studies; K. D. and J. A. conducted the unbiased screen; J. M. K. conducted the targeted screen; J. M. K. performed the mammalian cell culture experiments and wrote the manuscript; all authors contributed to editing the manuscript; A. C. T., C. D. S., M. O. H. and K. S. R. contributed to the data analysis, project planning and writing of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1, S2, S3, S4, S5, S6, Supplementary Tables S1, S2, S3, S4, S5 and Supplementary Methods (PDF 1741 kb)
Rights and permissions
About this article
Cite this article
Kinchen, J., Doukoumetzidis, K., Almendinger, J. et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol 10, 556–566 (2008). https://doi.org/10.1038/ncb1718
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1718
This article is cited by
-
Mesenchymal stem cell-derived apoptotic bodies alleviate alveolar bone destruction by regulating osteoclast differentiation and function
International Journal of Oral Science (2023)
-
Efferocytosis requires periphagosomal Ca2+-signaling and TRPM7-mediated electrical activity
Nature Communications (2022)
-
Circulating extracellular vesicles and rheumatoid arthritis: a proteomic analysis
Cellular and Molecular Life Sciences (2022)
-
SUMO modification in apoptosis
Journal of Molecular Histology (2021)
-
Hemocyte phagosomal proteome is dynamically shaped by cytoskeleton remodeling and interorganellar communication with endoplasmic reticulum during phagocytosis in a marine invertebrate, Crassostrea gigas
Scientific Reports (2020)