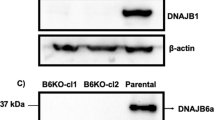
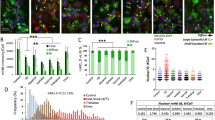
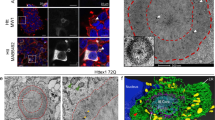
Abstract
Nuclear dysfunction is a key feature of the pathology of polyglutamine (polyQ) diseases. It has been suggested that mutant polyQ proteins impair functions of nuclear factors by interacting with them directly in the nucleus. However, a systematic analysis of quantitative changes in soluble nuclear proteins in neurons expressing mutant polyQ proteins has not been performed. Here, we perform a proteome analysis of soluble nuclear proteins prepared from neurons expressing huntingtin (Htt) or ataxin-1 (AT1) protein, and show that mutant AT1 and Htt similarly reduce the concentration of soluble high mobility group B1/2 (HMGB1/2) proteins. Immunoprecipitation and pulldown assays indicate that HMGBs interact with mutant AT1 and Htt. Immunohistochemistry showed that these proteins were reduced in the nuclear region outside of inclusion bodies in affected neurons. Compensatory expression of HMGBs ameliorated polyQ-induced pathology in primary neurons and in Drosophila polyQ models. Furthermore, HMGBs repressed genotoxic stress signals induced by mutant Htt or transcriptional repression. Thus, HMGBs may be critical regulators of polyQ disease pathology and could be targets for therapy development.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Gusella, J. F. & MacDonald, M. E. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nature Rev. Neurosci, 1, 109–115 (2000).
Zoghbi, H. Y. & Orr, H. T. Glutamine repeats and neurodegeneration. Annu. Rev. Neurosci. 23, 217–247 (2000).
Ross, C. A. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 35, 819–822 (2002).
Taylor, J. P., Hardy, J. & Fischbeck, K. H. Toxic proteins in neurodegenerative disease. Science 296, 1991–1995 (2002).
Bonini, N. M. Chaperoning brain degeneration. Proc. Natl Acad. Sci. USA 99, S16407–S16311 (2002).
Muchowski, P. J. & Wacker J. L . Modulation of neurodegeneration by molecular chaperones. Nature Rev. Neurosci. 6, 11–22 (2005).
Orr, H. T. Beyond the Qs in the polyglutamine diseases. Genes Dev. 15, 925–932 (2001).
Venkatraman, P., Wetzel, R., Tanaka, M., Nukina, N. & Goldberg AL . Eukaryotic proteasomes cannot digest polyglutamine sequences and release them during degradation of polyglutamine-containing proteins. Mol Cell. 14, 95–104 (2004).
Panov, A. V. et al. Early mitochondrial calcium defects in Huntington's disease are a direct effect of polyglutamines. Nature Neurosci. 5, 731–736 (2002).
Li, H., Li, S. H., Johnston, H., Shelbourne, P. F. & Li, X. J. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nature Genet. 25, 385–389 (2000).
Gauthier, L. R. et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 118, 127–138 (2004).
Sugars, K. L. & Rubinsztein, D. C. Transcriptional abnormalities in Huntington disease. Trends Genet. 19, 233–238 (2003).
Klement, I. A. et al. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 95, 41–53 (1998).
Saudou, F., Finkbeiner, S., Devys, D. & Greenberg, M. E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell 95, 55–66 (1998).
Steffan, J. S. et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 413, 739–743 (2001).
McCampbell, A. et al. Histone deacetylase inhibitors reduce polyglutamine toxicity. Proc. Natl Acad. Sci. USA 98, 15179–15184 (2001).
Hockly, E. et al. Suberoylanilide hydroxamic acid, a histone deacetylase inhibitor, ameliorates motor deficits in a mouse model of Huntington's disease. Proc. Natl Acad. Sci. USA 100, 2041–2046 (2003).
Katsuno, M. et al. Leuprorelin rescues polyglutamine-dependent phenotypes in a transgenic mouse model of spinal and bulbar muscular atrophy. Nature Med. 9, 768–773 (2003).
Dignam, J. D., Lebovitz, R. M. & Roeder, R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 (1983).
Agresti, A. & Bianchi, M. E. HMGB proteins and gene expression. Curr. Opin. Genet Develop. 13, 170–178 (2003).
Travers A. E . Priming the nucleosome: a role for HMGB proteins? EMBO Rep. 4, 131–136 (2003).
Scherzinger, E. et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell. 90, 549–558 (1997).
Perutz, M. F., Pope B. J., Owen, D., Wanker, E. E. & Scherzinger, E. Aggregation of proteins with expanded glutamine and alanine repeats of the glutamine-rich and asparagines-rich domains of Sup35 and of the amyloid β-peptide of amyloid plaques. Proc. Natl Acad. Sci. USA 99, 5596–5600 (2002).
Ross, C. A., Poirier, M. A., Wanker, E. E. & Amzel, M. Polyglutamine fibrilogenesis: The pathway unfolds. Proc. Natl Acad. Sci. USA 100, 1–3 (2003).
Tagawa, K. et al. Distinct aggregation and cell death patterns among different types of primary neurons induced by mutant huntingtin protein. J. Neurochem. 89, 974–987 (2004).
Hoshino, M., Tagawa, K., Okuda, T. & Okazawa H . General transcriptional repression by polyglutamine disease proteins is not directly linked to the presence of inclusion bodies. Biochem. Biophys. Res. Commun. 313, 110–116 (2004).
Fernandez-Funez, P. et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature 408, 101–106 (2000).
Jackson, G. R. et al. Polyglutamine-expanded human huntingtin transgenes induce degeneration of Drosophila photoreceptor neurons. Neuron 21, 633–642 (1998).
Hoshino, M. et al. Transcriptional repression induces a slowly progressive atypical neuronal death associated with changes of YAP isoforms and p73. J. Cell Biol. 172, 589–604 (2006).
Bae, B.-II . et al. p53 mediates cellular dysfunction and behavioral abnormalities in Huntington's disease. Neuron 47, 29–41 (2005).
Sancar, A., Lindsay-Boltz, L. A., Unsal-Kacmaz, K. & Linn, S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoint. Annu. Rev. Biochem. 73, 39–85 (2004).
Sengupta, S. & Harris, C. C. p53: traffic cop at the crossroads of DNA repair and recombination. Nature Rev. Mol. Cell Biol. 6, 44–55 (2005).
Ohndorf, U.-M., Rould, M. A., He, Q., Pabo, C. O. & Lippard, S. J. Basis for recognition of cisplatin-modified DNA by high mobility group proteins. Nature 399, 708–712 (1999).
Jayaraman, L. et al. High mobility group protein-1 (HMG-1) is a unique activator of p53. Genes Dev. 12, 462–472 (1998).
Zhang, Y. et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 (2005).
Giavara, S. et al. Yeast Nph6A/B and mammalian Hmgb1 facilitate the maintenance of genome stability. Curr. Biol. 15, 68–72 (2005).
Bianchi, M. E. & Beltrame, M. Flexing DNA: HMG-box proteins and their partners. Am. J. Hum. Genet. 63, 1573–1577 (1998).
Schaffar G . et al. Cellular toxicity of polyglutamine expansion proteins: mechanism of transcription factor deactivation. Mol. Cell 15, 95–105 (2004).
Zhai, W., Jeong, H., Cui, L., Krainc, D. & Tjian, R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell 123, 1241–1253 (2005).
Okazawa, H. et al. Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron 34, 701–713 (2002).
Arrasate, M., Mitra, S., Schweitzer, E. S. & Finkbeiner, S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 (2005).
Bonaldi, T. et al. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 22, 5551–5560 (2003).
de Chiara, C. et al. The AXH module: an independently folded domain common to ataxin-1 and HBP1. FEBS Lett. 551, 107–112 (2003).
Chen, Y. W., Allen, M. D., Veprintsev, D. B., Lowe, J. & Bycroft, M. The structure of the AXH domain of spinocerebellar ataxin-1. J. Biol. Chem. 279, 3758–3765 (2004).
Calogero, S. et al. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycemia in newborn mice. Nature Genet. 22, 276–280 (1999).
Okamoto, K., Okazawa, H., Okuda, A., Muramatsu, M. & Hamada, H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell 60, 461–472 (1990).
Watase, K. et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron 34, 905–919 (2002).
Hirai, H. & Launey, T. The regulatory connection between the activity of granule cell NMDA receptors and dendritic differentiation of cerebellar Purkinje cells. J. Neurosci. 20, 5217–5224 (2000).
Dura, J. M., Preat, T. & Tully, T. Identification of linotte, a new gene affecting learning and memory in Drosophila melanogaster. J. Neurogenet. 9, 1–14 (1993).
Rubin, G. M. & Spradling, A. C. Genetic transformation of Drosophila with transposable element vectors. Science. 218, 348–353 (1982).
Acknowledgements
This work was supported by grants to H.O. from the Japan Ministry of Education, Culture, Science, Sports and Technology (16650076), Japan Society for the Promotion of Science (JSPS; 16390249) and Japan Science and Technology Agency (JST; PRESTO). We thank M. Matsumoto and T. Miyashita (Tokyo Metropolitan Institute for Neuroscience, TMIN) for Drosophila experiments, T. Okuda and Shigeki Marubuchi (TMIN, Tokyo Medical and Dental University; TMDU) for primary culture, and T. Tajima for immunohistochemistry. We also thank H. Y. Zoghbi (Baylor College of Medicine) for sharing AT1 knock-in mouse samples and for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
This work was mainly performed by M.-L.Q. in collaboration with K.T, Y.E. and other co-authors. The project was planned by H.O.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures S1, S2, S3, S4, S5, S6, S7 and Supplementary Methods (PDF 1509 kb)
Rights and permissions
About this article
Cite this article
Qi, ML., Tagawa, K., Enokido, Y. et al. Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol 9, 402–414 (2007). https://doi.org/10.1038/ncb1553
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb1553
This article is cited by
-
PQBP5/NOL10 maintains and anchors the nucleolus under physiological and osmotic stress conditions
Nature Communications (2023)
-
HMGB1 is a Potential and Challenging Therapeutic Target for Parkinson’s Disease
Cellular and Molecular Neurobiology (2023)
-
HMGB1 signaling phosphorylates Ku70 and impairs DNA damage repair in Alzheimer’s disease pathology
Communications Biology (2021)
-
DNAzyme Cleavage of CAG Repeat RNA in Polyglutamine Diseases
Neurotherapeutics (2021)
-
Long noncoding RNA BS-DRL1 modulates the DNA damage response and genome stability by interacting with HMGB1 in neurons
Nature Communications (2021)