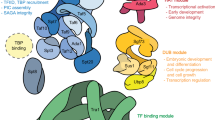
Abstract
A growing literature points to a fundamental role for the ubiquitin–proteasome degradation system (UPS) in transcription. Four recent publications add significant insight to our understanding of the connections between these processes. Each provides evidence that some aspect of the UPS can stimulate the activity of transcriptional activators. UPS might promote transcription by several mechanisms, and in some cases, even the final step of the UPS — proteolysis — might enhance activator function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
Change history
06 November 2003
'California Institute of Biology' changed to 'California Institute of Technology' in second author affiliation PDF has been REPLACED no erratum will be published
Notes
Institue of Biology has been amended to read Institute of Technology
References
Muratani, M. & Tansey, W.P. How the ubiquitin-proteasome system controls transcription. Nature Rev. Mol. Cell Biol. 4, 192–201 (2003).
Carr, A. Cell cycle: piecing together the p53 puzzle. Science 287, 1765 (2000).
Hart, M. et al. The F-box protein β-TrCP associates with phosphorylated β-catenin and regulates its activity in the cell. Curr. Biol. 9, 207–210 (1999).
Deroo, B.J. et al. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol. Cell. Biol. 22, 4113–4123 (2002).
Xie, Y. & Varshavsky, A. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc. Natl Acad. Sci. USA 98, 3056–3061 (2001).
Molinari, E., Gilman, M. & Natesan, S. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 18, 6439–6447 (1999).
Salghetti, S.E., Muratani, M., Wijnen, H., Futcher, B. & Tansey, W.P. Functional overlap of sequences that activate transcription and signal ubiquitin-mediated proteolysis. Proc. Natl Acad. Sci. USA 97, 3118–3123 (2000).
Ansari, A.Z. et al. Transcriptional activating regions target a cyclin-dependent kinase. Proc. Natl Acad. Sci. USA 99, 14706–14709 (2002).
Chi, Y. et al. Negative regulation of Gcn4 and Msn2 transcription factors by Srb10 cyclin-dependent kinase. Genes Dev. 15, 1078–1092 (2001).
Nelson, C., Goto, S., Lund, K., Hung, W. & Sadowski, I. Srb10/Cdk8 regulates yeast filamentous growth by phosphorylating the transcription factor Ste12. Nature 421, 187–190 (2003).
Salghetti, S.E., Caudy, A.A., Chenoweth, J.G. & Tansey, W.P. Regulation of transcriptional activation domain function by ubiquitin. Science 293, 1651–1653 (2001).
Braun, B. et al. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nature Cell Biol. 1, 221–226 (1999).
Gonzalez, F., Delahodde, A., Kodadek, T. & Johnston, S.A. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science 296, 548–550 (2002).
Reid, G. et al. Cyclic, proteasome-mediated turnover of unliganded and liganded ERα on responsive promoters is an integral feature of estrogen signaling. Mol. Cell 11, 695–707 (2003).
von der Lehr, N. et al. The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell 11, 1177–1188 (2003).
Kim, S., Herbst, A., Tworkowski, K., Salghetti, S. & Tansey, W. Skp2 regulates Myc protein stability and activity. Mol. Cell 11, 1177–1188 (2003).
Morris, M.C. et al. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 423, 1009–1013 (2003).
Lonard, D.M., Nawaz, Z., Smith, C.L. & O'Malley, B.W. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol. Cell 5, 939–948 (2000).
Lin, H.K. et al. Proteasome activity is required for androgen receptor transcriptional activity via regulation of androgen receptor nuclear translocation and interaction with coregulators in prostate cancer cells. J. Biol. Chem. 277, 36570–36576 (2002).
Stenoien, D.L. et al. FRAP reveals that mobility of oestrogen receptor-α is ligand- and proteasome-dependent. Nature Cell Biol. 3, 15–23 (2001).
Johnson, E.S., Gonda, D.K. & Varshavsky, A. Cis–trans recognition and subunit-specific degradation of short-lived proteins. Nature 346:287–291 (1990).
Verma, R., McDonald, W., Yates, J. & Deshaies, R. Selective degradation of ubiquitinated Sic1 by purified 26S proteasome yields active S phase cyclin-Cdk. Mol. Cell 8, 439–448 (2001).
Nishiyama, A. et al. A nonproteolytic function of the proteasome is required for the dissociation of Cdc2 and cyclin B at the end of M phase. Genes Dev. 14, 2344–2357 (2000).
Aparicio, O.M., Weinstein, D.M. & Bell, S.P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69 (1997).
Kominami, K. et al. Nin1p, a regulatory subunit of the 26S proteasome, is necessary for activation of Cdc28p kinase of Saccharomyces cerevisiae. EMBO J. 14, 3105–3115 (1995).
Tansey, W.P. Transcriptional activation: risky business. Genes Dev. 15, 1045–1050 (2001).
Natarajan, K., Jackson, B.M., Zhou, H., Winston, F. & Hinnebusch, A.G. Transcriptional activation by Gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol. Cell 4, 657–664 (1999).
Freeman, B.C. & Yamamoto, K.R. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science 296, 2232–2235 (2002).
Thomas, D. & Tyers, M. Transcriptional regulation: kamikaze activators. Curr. Biol. 10, R341–R343 (2000).
Tworkowski, K.A., Salghetti, S.E. & Tansey, W.P. Stable and unstable pools of Myc protein exist in human cells. Oncogene 21, 8515–8520 (2002).
Kuras, L. et al. Dual regulation of the met4 transcription factor by ubiquitin-dependent degradation and inhibition of promoter recruitment. Mol. Cell 10, 69–80 (2002).
Freiman, R.N. & Tijian, R. Regulating the regulators: lysine modifications make their mark. Cell 112, 11–17 (2003).
Hershko, A. & Ciechanover, A. The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 (1998).
Glickman, M. et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell 94, 615–623 (1998).
Verma, R. et al. Deubiquitination and degradation of proteins by the 26S proteasome requires the Rpn11 metalloprotease motif. Science 298, 611–615 (2002).
Yao, T. & Cohen, R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419, 403–407 (2002).
Musti, A.M., Treier, M. & Bohmann, D. Reduced ubiquitin-dependent degradation of c-Jun after phosphorylation by MAP kinases. Science 275, 400–402 (1997).
Kim, W. & Kaelin, W.J. The von Hippel–Lindau tumor suppressor protein: new insights into oxygen sensing and cancer. Curr. Opin. Genet. Dev. 13, 55–60 (2003).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lipford, J., Deshaies, R. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat Cell Biol 5, 845–850 (2003). https://doi.org/10.1038/ncb1003-845
Issue Date:
DOI: https://doi.org/10.1038/ncb1003-845
This article is cited by
-
RCB initiates Arabidopsis thermomorphogenesis by stabilizing the thermoregulator PIF4 in the daytime
Nature Communications (2021)
-
Transcriptional regulatory proteins in central carbon metabolism of Pichia pastoris and Saccharomyces cerevisiae
Applied Microbiology and Biotechnology (2020)
-
E3 ubiquitin ligases in cancer and implications for therapies
Cancer and Metastasis Reviews (2017)
-
Root development: Pulse control
Nature Plants (2015)
-
dDsk2 regulates H2Bub1 and RNA polymerase II pausing at dHP1c complex target genes
Nature Communications (2015)