Abstract
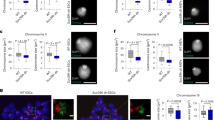
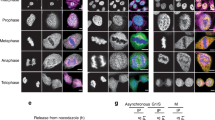
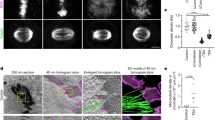
Histone modifications might act to mark and maintain functional chromatin domains during both interphase and mitosis. Here we show that pericentric heterochromatin in mammalian cells is specifically responsive to prolonged treatment with deacetylase inhibitors. These defined regions relocate at the nuclear periphery and lose their properties of retaining HP1 (heterochromatin protein 1) proteins. Subsequent defects in chromosome segregation arise in mitosis. All these changes can reverse rapidly after drug removal. Our data point to a crucial role of histone underacetylation within pericentric heterochromatin regions for their association with HP1 proteins, their nuclear compartmentalization and their contribution to centromere function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Lamond, A. I. & Earnshaw, W. C. Structure and function in the nucleus. Science 280, 547– 553 (1998).
Cockell, M. & Gasser, S. M. Nuclear compartments and gene regulation. Curr. Opin. Genet. Dev. 9, 199 –205 (1999).
Dobie, K. W., Hari, K. L., Maggert, K. A. & Karpen, G. H. Centromere proteins and chromosome inheritance: a complex affair. Curr. Opin. Genet. Dev. 9, 206–217 (1999).
Craig, J. M., Earnshaw, W. C. & Vagnarelli, P. Mammalian centromeres: DNA sequence, protein composition, and role in cell cycle progression. Exp. Cell Res. 246, 249–262 (1999).
Eissenberg, J. C. & Elgin, S. C. The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204–210 (2000).
Jones, D. O., Cowel, I. G. & Singh, P. B. Mammalian chromodomain proteins: their role in genome organisation and expression. Bioessays 22, 124–137 (2000).
Wakimoto, B. T. Beyond the nucleosome: epigenetic aspects of position–effect variegation in Drosophila. Cell 93, 321– 324 (1998).
Festenstein, R. et al. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nature Genet. 23, 457–461 (1999).
Ekwall, K. et al. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269, 1429– 1431 (1995).
Kellum, R. & Alberts, B. M. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 108, 1419–1431 (1995).
Turner, B. M. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell. Mol. Life Sci. 54, 21– 31 (1998).
Strahl, B. D. & Allis, C. D. The language of covalent histone modifications. Nature 403, 41– 45 (2000).
Grunstein, M. Yeast heterochromatin: regulation of its assembly and inheritance by histones . Cell 93, 325–328 (1998).
Ekwall, K., Olsson, T., Turner, B. M., Cranston, G. & Allshire, R. C. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91, 1021– 1032 (1997).
Grewal, S. I., Bonaduce, M. J. & Klar, A. J. Histone deacetylase homologs regulate epigenetic inheritance of transcriptional silencing and chromosome segregation in fission yeast. Genetics 150, 563– 576 (1998).
Taddei, A., Roche, D., Sibarita, J.B., Turner, B. M. & Almouzni, G. Duplication and maintenance of heterochromatin domains. J. Cell Biol. 147, 1153–1166 (1999).
Rea, S. et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases . Nature 406, 593–599 (2000).
Paro, R. Chromatin regulation. Formatting genetic text. Nature 406, 579–580 (2000).
Yoshida, M., Horinouchi, S. & Beppu, T. Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. Bioessays 17, 423–430 ( 1995).
Cerda, M. C., Berrios, S., Fernandez-Donoso, R., Garagna, S. & Redi, C. Organisation of complex nuclear domains in somatic mouse cells. Biol. Cell 91, 55 –65 (1999).
Bird, A. The essentials of DNA methylation. Cell 70, 5–8 (1992).
Habib, M. et al. DNA global hypomethylation in EBV-transformed interphase nuclei . Exp. Cell Res. 249, 46– 53 (1999).
Earnshaw, W. C. & Rothfield, N. Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91, 313– 321 (1985).
Belyaev, N. D., Keohane, A. M. & Turner, B. M. Histone H4 acetylation and replication timing in Chinese hamster chromosomes. Exp. Cell Res. 225, 277–285 (1996).
Misteli, T. & Spector, D. L. The cellular organization of gene expression. Curr. Opin. Cell Biol. 10, 323–331 (1998).
Puvion-Dutilleul, F. et al. Alterations of nucleolar ultrastructure and ribosome biogenesis by actinomycin D. Implications for U3 snRNP function. Eur. J. Cell Biol. 58, 149–162 ( 1992).
Marheineke, K. & Krude, T. Nucleosome assembly activity and intracellular localization of human CAF-1 changes during the cell division cycle. J. Biol. Chem. 273, 15279–15286 (1998).
Martini, E, Roche, D. M., Marheineke, K., Verreault, A. & Almouzni, G. Recruitment of phosphorylated chromatin assembly factor 1 to chromatin after UV irradiation of human cells. J. Cell Biol. 143, 563–575 (1998).
Remboutsika, E. et al. The putative nuclear receptor mediator TIF1α is tightly associated with euchromatin. J. Cell Sci. 112, 1671–1683 (1999).
Nielsen, A. L. et al. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 18 , 6385–6395 (1999).
Dini, L., Coppola, S., Ruzittu, M. T. & Ghibelli, L. Multiple pathways for apoptotic nuclear fragmentation. Exp. Cell Res. 223, 340–347 ( 1996).
Marrazzini, A., Betti, C., Bernacchi, F., Barrai, I. & Barale, R. Micronucleus test and metaphase analyses in mice exposed to known and suspected spindle poisons. Mutagenesis 9, 505–515 (1994).
Shimizu, N., Itoh, N., Utiyama, H. & Wahl, G. M. Selective entrapment of extrachromosomally amplified DNA by nuclear budding and micronucleation during S phase. J. Cell Biol. 140, 1307– 1320 (1998).
Sogo, J. M. & Laskey, R. A. in Chromatin Structure and Gene Expression (ed. Elgin, S. C. R.) 49–71 (Oxford Univ. Press, New York, 1995).
Zhao, T., Heyduk, T., Allis, C. D. & Eissenberg, J. C. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 275, 28332–28338 ( 2000).
Knoepfler, P. S. & Eisenman, R. N. Sin meets NuRD and other tails of repression. Cell 99, 447–450 (1999).
Robertson, K. D. et al. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet. 25, 338–342 (2000).
Rountree, M. R., Bachman, K. E. & Baylin, S. B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet. 25, 269–277 (2000).
Xu, G. L. et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature 402 , 187–191 (1999).
Turner, B. M. & Fellows, G. Specific antibodies reveal ordered and cell-cycle-related use of histone-H4 acetylation sites in mammalian cells . Eur. J. Biochem. 179, 131– 139 (1989).
Maison, C., Pyrpasopoulou, A. & Georgatos, S. D. Vimentin-associated mitotic vesicles interact with chromosomes in a lamin B- and phosphorylation-dependent manner. EMBO J. 14, 3311–3324 ( 1995).
Acknowledgements
We thank P. Chambon, B. Turner, C. Marheineke and A. Niveleau for providing antibodies, W. Earnshaw for the HP1–GST clone, ACA and anti-CENP-C antibodies, and J. Mello for critical reading. A.T. was supported by fellowships from the Ministère de l'Éducation Nationale et de l'Enseignement Supérieur de la Recherche et de la Technologie, l'Association de la Recherche sur le Cancer and the Human Frontier Science Foundation. This work was supported by EU TMR and by Ligue National contre le Cancer.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Figure S1 DNA-synthesis inhibitors do not affect the localization of centromeres.
Figure S2 Three different inhibitors of histone deacetylases affect pericentric regions similarly to TSA. (PDF 197 kb)
Figure S3 Inhibition of DNA methylation with 5-azacytidine does not affect the association of HP1 with pericentric regions.
Supplementary references.
Rights and permissions
About this article
Cite this article
Taddei, A., Maison, C., Roche, D. et al. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol 3, 114–120 (2001). https://doi.org/10.1038/35055010
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/35055010
This article is cited by
-
SETDB1-like MET-2 promotes transcriptional silencing and development independently of its H3K9me-associated catalytic activity
Nature Structural & Molecular Biology (2022)
-
Satellite repeat transcripts modulate heterochromatin condensates and safeguard chromosome stability in mouse embryonic stem cells
Nature Communications (2022)
-
Microbiota derived short chain fatty acids promote histone crotonylation in the colon through histone deacetylases
Nature Communications (2018)
-
Vorinostat differentially alters 3D nuclear structure of cancer and non-cancerous esophageal cells
Scientific Reports (2016)
-
Human centromere repositioning within euchromatin after partial chromosome deletion
Chromosome Research (2016)