Abstract
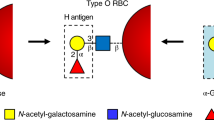
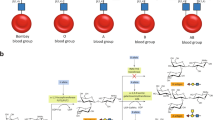
Enzymatic removal of blood group ABO antigens to develop universal red blood cells (RBCs) was a pioneering vision originally proposed more than 25 years ago. Although the feasibility of this approach was demonstrated in clinical trials for group B RBCs, a major obstacle in translating this technology to clinical practice has been the lack of efficient glycosidase enzymes. Here we report two bacterial glycosidase gene families that provide enzymes capable of efficient removal of A and B antigens at neutral pH with low consumption of recombinant enzymes. The crystal structure of a member of the α-N-acetylgalactosaminidase family reveals an unusual catalytic mechanism involving NAD+. The enzymatic conversion processes we describe hold promise for achieving the goal of producing universal RBCs, which would improve the blood supply while enhancing the safety of clinical transfusions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Landsteiner, K. Agglutination phenomena of normal human blood. Wien. Klin. Wochenschr. 113, 768–769 (2001).
Watkins, W.M. Biochemistry and Genetics of the ABO, Lewis, and P blood group systems. Adv. Hum. Genet. 10, 1–136, 379–185 (1980).
Clausen, H. & Hakomori, S. ABH and related histo-blood group antigens; immunochemical differences in carrier isotypes and their distribution. Vox Sang. 56, 1–20 (1989).
Yamamoto, F., Clausen, H., White, T., Marken, J. & Hakomori, S. Molecular genetic basis of the histo-blood group ABO system. Nature 345, 229–233 (1990).
Sazama, K. Transfusion errors: scope of the problem, consequences, and solutions. Curr. Hematol. Rep. 2, 518–521 (2003).
Stainsby, D. et al. Serious Hazards of Transfusion Annual Report 2004 (Serious Hazards of Transfusion Office, Manchester Blood Centre, Manchester, UK; 2005).
Olsson, M.L. et al. Universal red blood cells–enzymatic conversion of blood group A and B antigens. Transfus. Clin. Biol. 11, 33–39 (2004).
Goldstein, J., Siviglia, G., Hurst, R., Lenny, L. & Reich, L. Group B erythrocytes enzymatically converted to group O survive normally in A, B, and O individuals. Science 215, 168–170 (1982).
Kruskall, M.S. et al. Transfusion to blood group A and O patients of group B RBCs that have been enzymatically converted to group O. Transfusion 40, 1290–1298 (2000).
Vosnidou, N.C. et al. Seroconversion of type B to O erythrocytes using recombinant Glycine max α-D-galactosidase. Biochem. Mol. Biol. Int. 46, 175–186 (1998).
Bakunina, I.Y. et al. Alpha-galactosidase of the marine bacterium Pseudoalteromonas sp. KMM 701. Biochemistry (Mosc.) 63, 1209–1215 (1998).
Clausen, H., Levery, S.B., Kannagi, R. & Hakomori, S. Novel blood group H glycolipid antigens exclusively expressed in blood group A and AB erythrocytes (type 3 chain H). I. Isolation and chemical characterization. J. Biol. Chem. 261, 1380–1387 (1986).
Clausen, H., Levery, S.B., Nudelman, E., Tsuchiya, S. & Hakomori, S. Repetitive A epitope (type 3 chain A) defined by blood group A1-specific monoclonal antibody TH-1: chemical basis of qualitative A1 and A2 distinction. Proc. Natl. Acad. Sci. USA 82, 1199–1203 (1985).
Zhu, A., Monahan, C., Wang, Z.K. & Goldstein, J. Expression, purification, and characterization of recombinant α-N-acetylgalactosaminidase produced in the yeast Pichia pastoris. Protein Expr. Purif. 8, 456–462 (1996).
Hsin-Yeh, H., Chapman, L.F., Calcutt, M.J. & Smith, D.S. Recombinant Clostridium perfringens α-N-acetylgalactosaminidase blood group A2 degrading activity. Artif. Cells Blood Substit. Immobil. Biotechnol. 33, 187–199 (2005).
Bakunina, I.Y. et al. α-N-acetylgalactosaminidase from marine bacterium Arenibacter latericius KMM 426T removing blood type specificity of A-erythrocytes. Biochemistry (Mosc.) 67, 689–695 (2002).
Landry, D. Isolation and composition of a novel glycosidase from chryseobacterium. US patent 6,458,525. (2002).
Pikis, A., Immel, S., Robrish, S.A. & Thompson, J. Metabolism of sucrose and its five isomers by Fusobacterium mortiferum. Microbiology 148, 843–852 (2002).
Yip, V.L. et al. An unusual mechanism of glycoside hydrolysis involving redox and elimination steps by a family 4 beta-glycosidase from Thermotoga maritima. J. Am. Chem. Soc. 126, 8354–8355 (2004).
Rajan, S.S. et al. Novel catalytic mechanism of glycoside hydrolysis based on the structure of an NAD+/Mn2+-dependent phospho-alpha-glucosidase from Bacillus subtilis. Structure 12, 1619–1629 (2004).
Yip, V.L. & Withers, S.G. Mechanistic analysis of the unusual redox-elimination sequence employed by Thermotoga maritima bglt: a 6-phospho-beta-glucosidase from glycoside hydrolase family 4. Biochemistry 45, 571–580 (2006).
Lesk, A.M. NAD-binding domains of dehydrogenases. Curr. Opin. Struct. Biol. 5, 775–783 (1995).
Holm, L. & Sander, C. Dali: a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 (1995).
Kingston, R.L., Scopes, R.K. & Baker, E.N. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure 4, 1413–1428 (1996).
Varrot, A. et al. NAD+ and metal-ion dependent hydrolysis by family 4 glycosidases: structural insight into specificity for phospho-beta-D-glucosides. J. Mol. Biol. 346, 423–435 (2005).
Yip, V.L. & Withers, S.G. Family 4 glycosidases carry out efficient hydrolysis of thioglycosides by an alpha,beta-elimination mechanism. Angew. Chem. Int. Edn. Engl. 45, 6179–6182 (2006).
Ishikura, H., Arakawa, S., Nakajima, T., Tsuchida, N. & Ishikawa, I. Cloning of the Tannerella forsythensis (Bacteroides forsythus) siaHI gene and purification of the sialidase enzyme. J. Med. Microbiol. 52, 1101–1107 (2003).
Goldstein, J., Lenny, L., Davies, D. & Voak, D. Further evidence for the presence of A antigen on group B erythrocytes through the use of specific exoglycosidases. Vox Sang. 57, 142–146 (1989).
Clausen, H., Holmes, E. & Hakomori, S. Novel blood group H glycolipid antigens exclusively expressed in blood group A and AB erythrocytes (type 3 chain H). II. Differential conversion of different H substrates by A1 and A2 enzymes, and type 3 chain H expression in relation to secretor status. J. Biol. Chem. 261, 1388–1392 (1986).
Davies, G.J., Gloster, T.M. & Henrissat, B. Recent structural insights into the expanding world of carbohydrate-active enzymes. Curr. Opin. Struct. Biol. 15, 637–645 (2005).
Henrissat, B. & Davies, G.J. Glycoside hydrolases and glycosyltransferases. Families, modules, and implications for genomics. Plant Physiol. 124, 1515–1519 (2000).
Zhu, A. et al. Characterization of recombinant alpha-galactosidase for use in seroconversion from blood group B to O of human erythrocytes. Arch. Biochem. Biophys. 327, 324–329 (1996).
Davis, M.O., Hata, D.J., Johnson, S.A., Walker, J.C. & Smith, D.S. Cloning, expression and characterization of a blood group B active recombinant α-D-galactosidase from soybean (Glycine max). Biochem. Mol. Biol. Int. 39, 471–485 (1996).
Calcutt, M.J., Hsieh, H.Y., Chapman, L.F. & Smith, D.S. Identification, molecular cloning and expression of an α-N-acetylgalactosaminidase gene from Clostridium perfringens. FEMS Microbiol. Lett. 214, 77–80 (2002).
Henrissat, B. Glycosidase families. Biochem. Soc. Trans. 26, 153–156 (1998).
Rye, C.S. & Withers, S.G. Glycosidase mechanisms. Curr. Opin. Chem. Biol. 4, 573–580 (2000).
Leslie, A.G.W. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26 (1992).
CCP4. The CCP4 Suite. Programs for protein crystallography. Acta Crystallogr D Biol. Crystallogr. 50, 760–763 (1994).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58, 1772–1779 (2002).
Sheldrick, G.M. Macromolecular phasing with SHELXE. Z. Kristallogr. 217, 644–650 (2002).
Perrakis, A., Morris, R. & Lamzin, V.S. Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 (1999).
Roussel, A. & Cambillau, C. TURBO-FRODO. in Silicon Graphics Geometry Partners Directory (eds. Silicon Graphics Committee) 77–78 (Silicon Graphics, Mountain View, California, 1989).
Murshudov, G.N., Vagin, A.A. & Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53, 240–255 (1997).
Hooft, R.W., Vriend, G., Sander, C. & Abola, E.E. Errors in protein structures. Nature 381, 272 (1996).
Technical Manual (AABB, Bethesda, Maryland, USA; 2005).
Clausen, H., Levery, S.B., McKibbin, J.M. & Hakomori, S. Blood group A determinants with mono- and difucosyl type 1 chain in human erythrocyte membranes. Biochemistry 24, 3578–3586 (1985).
Acknowledgements
This paper is dedicated to the memory of Margot Kruskall. We thank Phil Robbins for his invaluable help throughout this work. We are grateful to Annika Hult at the Blood Center in Lund for technical assistance with FACS analysis and to Etienne Danchin, AFMB, for preparing Supplementary Fig. 1 online. This work was supported by ZymeQuest Inc. and the Centre National de la Recherche Scientifique. Work performed in M.L.O.'s laboratory was supported by the Swedish Research Council (project no. K2005-71X-14251), governmental ALF research grants to Lund University Hospital, the Inga and John Hain Foundation for Medical Research and Region Skåne, Sweden. The European Synchrotron Radiation Facility (ESRF) is acknowledged for beam time allocation.
Author information
Authors and Affiliations
Contributions
Q.P.L. contributed to screening and purification of enzymes, planning and design of the project, and manuscript writing; G.S. contributed to the X-ray crystallography and manuscript writing; H.Y. contributed to purification of enzymes; E.P.B. and K.S. contributed to cloning of genes; G.P. contributed to cloning of genes; J.S. and K.S. contributed to development of enzyme conversion protocol; E.N. contributed to glycolipid analysis; S.B.L. contributed with NMR analysis; T.W. contributed to planning and design of the project; J.M.N. and W.S.L. contributed to sequencing of purified enzyme; Y.B. contributed to manuscript writing; M.L.O. contributed to FACS analysis, planning and design of the project and manuscript writing; B.H. contributed to planning and manuscript writing; H.C. contributed to planning and design of the project, and manuscript writing.
Corresponding authors
Ethics declarations
Competing interests
Authors (except for G.S., J.M.N., W.S.L. and Y.V.) are employees, consultants and/or shareholders in Zymequest Inc., which holds patents covering the described technologies.
Supplementary information
Supplementary Fig. 1
Maximum Likelihood phylogenetic tree of the GH109 α-N-acetylgalactosaminidase (a) and the GH110 α-galactosidase (b) families. (PDF 131 kb)
Supplementary Fig. 2
Influence of different additives on the recombinant E. meningosepticum α-N-acetylgalactosaminidase. (PDF 94 kb)
Supplementary Fig. 3
Analysis of pH optimum of E. meningosepticum α-N-acetylgalactosaminidase and FragA α-galactosidase using AMC-labeled tetrasaccharides by TLC. (PDF 147 kb)
Supplementary Fig. 4
Substrate specificities of E. meningosepticum α-N-acetylgalactosaminidase and FragA α-galactosidase. (PDF 142 kb)
Supplementary Fig. 5
Analysis of hydrolysis of GalNAcβ-pNP by the E. meningosepticum α-N-acetylgalactosaminidase. (PDF 175 kb)
Supplementary Fig. 6
The E. meningosepticum α-N-acetylgalactosaminidase is dependent on NAD+ as cofactor. (PDF 130 kb)
Supplementary Fig. 7
NAD+ dependence of E. meningosepticum α-N-acetylgalactosaminidase activity with monosaccharide pNP substrates. (PDF 193 kb)
Supplementary Fig. 8
Cartoon representation of the active site of the E. meningosepticum α-N-acetylgalactosaminidase. (PDF 35 kb)
Supplementary Fig. 9
Multiple sequence alignment analysis of novel glycosidase families GH109 and 110. (PDF 638 kb)
Supplementary Fig. 10
Proposed mechanism of the E. meningosepticum α-N-acetylgalactosaminidase. (PDF 137 kb)
Supplementary Table 1
Data collection and refinement statistics (PDF 22 kb)
Rights and permissions
About this article
Cite this article
Liu, Q., Sulzenbacher, G., Yuan, H. et al. Bacterial glycosidases for the production of universal red blood cells. Nat Biotechnol 25, 454–464 (2007). https://doi.org/10.1038/nbt1298
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1298
This article is cited by
-
Enhancement of α-galactosidase production using novel Actinoplanes utahensis B1 strain: sequential optimization and purification of enzyme
World Journal of Microbiology and Biotechnology (2024)
-
Host genetic regulation of human gut microbial structural variation
Nature (2024)
-
Turning universal O into rare Bombay type blood
Nature Communications (2023)
-
Enzymatic β-elimination in natural product O- and C-glycoside deglycosylation
Nature Communications (2023)
-
Theoretical study on the glycosidic C–C bond cleavage of 3’’-oxo-puerarin
Scientific Reports (2023)