Abstract
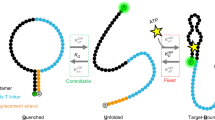
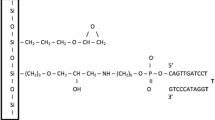
Reagentless biosensors that can directly transduce molecular recognition to optical signals should potentiate the development of sensor arrays for a wide variety of analytes. Nucleic acid aptamers that bind ligands tightly and specifically can be readily selected, but may prove difficult to adapt to biosensor applications. We have therefore attempted to develop selection methods that couple the broad molecular recognition properties of aptamers with signal transduction. Anti-adenosine aptamers were selected from a pool that was skewed to contain very few fluoresceinated uridines. The primary family of aptamers showed a doubling of relative fluorescence intensity at saturating concentrations of a cognate analyte, ATP, and could sense ATP concentrations as low as 25 μM. A single uridine was present in the best signaling aptamer. Surprisingly, other dyes could substitute for fluorescein and still specifically signal the presence of ATP, indicating that the single uridine functioned as a general “switch” for transducing molecular recognition to optical signals.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Hesselberth, J., Robertson, M.P., Jhaveri, S.D. & Ellington, A.D. In vitro selection of nucleic acids for diagnostic applications. Rev. Mol. Biotechnol. 74, 15–25 (2000).
Jayasena,S.D. Aptamers: an emerging class of molecules that rival antibodies in diagnostics . Clin. Chem. 45, 1628– 1650 (1999)
Famulok, M. & Jenne, A. Oligonucleotide libraries—variatio delectat. Curr. Opin. Chem. Biol. 2, 320 –327 (1998).
Gold, L., Polisky, B., Uhlenbeck, O. & Yarus, M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 64, 763–797 (1995).
Osborne, S.E., Matsumura, I. & Ellington, A.D. Aptamers as therapeutic and diagnostic reagents: problems and prospects. Curr. Opin. Chem. Biol. 1, 5–9 (1997).
Famulok, M. & Mayer, G. Aptamers as tools in molecular biology and immunology. Curr. Top. Microbiol. Immunol. 243, 123–136 ( 1999).
Geiger, A., Burgstaller, P., von der Eltz, H., Roeder, A. & Famulok, M. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 24, 1029–1036 (1996).
Morris, K.N., Jensen, K.B., Julin, C.M., Weil, M. & Gold, L. High affinity ligands from in vitro selection: complex targets. Proc. Natl. Acad. Sci. USA 95, 2902–2907 (1998).
German, I., Buchanan, D.D. & Kennedy, R.T. Aptamers as ligands in affinity probe capillary electrophoresis . Anal. Chem. 70, 4540– 4545 (1998).
Potyrailo, R.A., Conrad, R.C., Ellington, A.D. & Hieftje, G.M. Adapting selected nucleic acid ligands (aptamers) to biosensors . Anal. Chem. 70, 3419– 3425 (1998).
Davis, K.A., Lin, Y., Abrams, B. & Jayasena, S.D. Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res. 26, 3915–3924 (1998).
Davis, K.A., Abrams, B., Lin, Y. & Jayasena, S.D. Use of a high affinity DNA ligand in flow cytometry. Nucleic Acids Res. 24, 702–706 (1996).
Giuliano, K.A. & Taylor, D.L. Fluorescent-protein biosensors: new tools for drug discovery. Trends Biotechnol. 16, 135–140 (1998).
Hirshberg, M. et al. Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate. Biochemistry 37, 10381–10385 (1998).
Brune, M. et al. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli. Biochemistry 37, 10370–10380 (1998).
Gilardi, G., Zhou, L.Q., Hibbert, L. & Cass, A.E. Engineering the maltose binding protein for reagentless fluorescence sensing. Anal. Chem. 66, 3840–3847 (1994).
Marvin, J.S. et al. The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc. Natl. Acad. Sci. USA 94, 4366–4371 (1997).
Adams, S.R., Harootunian, A.T., Buechler, Y.J., Taylor, S.S. & Tsien, R.Y. Fluorescence ratio imaging of cyclic AMP in single cells. Nature 349, 694–697 (1991).
Patel, D.J. et al. Structure, recognition and adaptive binding in RNA aptamer complexes . J. Mol. Biol. 272, 645– 664 (1997).
Westhof, E. & Patel, D.J. Nucleic acids. From self-assembly to induced-fit recognition. Curr. Opin. Struct. Biol. 7, 305–309 ( 1997).
Jhaveri, S.D. et al. Designed signaling aptamers that transduce molecular recognition to changes in fluorescence intensity. J. Am. Chem. Soc. 122, 2469–2473 (2000)
Rogers, J. & Joyce, G.F. A ribozyme that lacks cytidine. Nature 402, 323– 325 (1999).
Sassanfar, M. & Szostak, J.W. An RNA motif that binds ATP. Nature 364, 550– 553 (1993).
Crick, F.H. The origin of the genetic code. J. Mol. Biol. 38, 367–379 (1968).
Wachtershauser, G. An all-purine precursor of nucleic acids. Proc. Natl. Acad. Sci. USA 85, 1134–1135 ( 1988).
Burke, D.H. & Gold, L. RNA aptamers to the adenosine moiety of S-adenosyl methionine: structural inferences from variations on a theme and the reproducibility of SELEX. Nucleic Acids Res. 25, 2020–2024 ( 1997).
Burgstaller, P. & Famulok, M. Isolation of RNA-aptamers for biological cofactors by in vitro selection. Angew. Chem. Int. Edn. Engl. 33, 1084–1087 (1994)
Ellington, A.D., Hesselberth, J., Jhaveri, S. & Robertson, M.P. Combinatorial methods: aptamers and aptazymes Proc. SPIE 3858, 126–134 (1999)
Taylor, L.C. & Walt, D.R. Application of high density optical microwell arrays in a live-cell biosensing system Anal. Biochem. 278, 132–142 (2000)
Walt, D.R. Techview: molecular biology. Bead-based fiber-optic arrays Science 287, 451–452 ( 2000).
Savoy, S. et al. Solution based analysis of multiple analytes by a sensor array: toward the development of an “electronic tongue.” Proc. SPIE 3539, 17–26 (1998)
Lavigne, J.J. et al. Single-analyte to multianalyte fluorescence sensors Proc. SPIE 3602, 220–231 ( 1999).
Pollard, J., Bell, S. & Ellington, A. Design, synthesis, and amplification of DNA pools for in vitro selection, In Current protocols in nucleic acid chemistry (ed. Beaucage, S.) 9.2.1–9.2.23 (Wiley, New York, NY; 2000).
Gallagher, S. Quantitation of DNA and RNA with absorption and fluorescence spectroscopy In Current protocols in molecular biology. (ed. Ausubel, F.) A.3D.1-A.3D.8 (Wiley, New York, NY; 2000 ).
Acknowledgements
This work was supported by a grant from Foundation for Research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jhaveri, S., Rajendran, M. & Ellington, A. In vitro selection of signaling aptamers. Nat Biotechnol 18, 1293–1297 (2000). https://doi.org/10.1038/82414
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/82414
This article is cited by
-
Split aptamer-based detection of adenosine triphosphate using surface enhanced Raman spectroscopy and two kinds of gold nanoparticles
Microchimica Acta (2019)
-
Colorimetric thrombin assay using aptamer-functionalized gold nanoparticles acting as a peroxidase mimetic
Microchimica Acta (2016)
-
Cell-targeting aptamers act as intracellular delivery vehicles
Applied Microbiology and Biotechnology (2016)
-
In vitro selection of a peptide aptamer that changes fluorescence in response to verotoxin
Biotechnology Letters (2015)
-
Aptamer-based molecular imaging
Protein & Cell (2012)