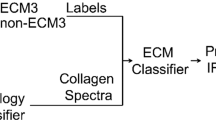
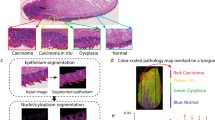
Abstract
The process of histopathology, comprising tissue staining and morphological pattern recognition, has remained largely unchanged for over 140 years1. Although it is integral to clinical and research activities, histopathologic recognition remains a time-consuming, subjective process to which only limited statistical confidence can be assigned because of inherent operator variability2,3. Although immunohistochemical approaches allow limited molecular detection, significant challenges remain in using them for quantitative, automated pathology. Vibrational spectroscopic approaches, by contrast, directly provide nonperturbing molecular descriptors4, but a practical spectroscopic protocol for histopathology is lacking. Here we couple high-throughput Fourier transform infrared (FTIR) spectroscopic imaging5 of tissue microarrays6 with statistical pattern recognition of spectra indicative of endogenous molecular composition and demonstrate histopathologic characterization of prostatic tissue. This automated histologic segmentation is applied to routine archival tissue samples, incorporates well-defined tests of statistical significance7 and eliminates any requirement for dyes or molecular probes. Finally, we differentiate benign from malignant prostatic epithelium by spectroscopic analyses.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Luna, L.G. Manual of Histologic Staining Methods of the Armed Forces Institute of Pathology (McGraw-Hill, New York, 1968).
Allsbrook, W.C., Jr. et al. Interobserver reproducibility of Gleason grading of prostatic carcinoma: urologic pathologists. Hum. Pathol. 32, 74–80 (2001).
Emmert-Buck, M.R. et al. Molecular profiling of clinical tissue specimens—feasibility and applications. Am. J. Pathol. 156, 1109–1115 (2000).
Diem, M. et al. A decade of vibrational micro-spectroscopy of human cells and tissue (1994–2004). Analyst 129, 880–885 (2004).
Lewis, E.N. et al. Fourier-transform spectroscopic imaging using an infrared focal-plane array detector. Anal. Chem. 67, 3377–3381 (1995).
Kononen, J. et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat. Med. 4, 844–847 (1998).
Baak, J.P.A. The framework of pathology: Good Laboratory Practice by quantitative and molecular methods. J. Pathol. 198, 277–283 (2002).
Jackson, M. & Mantsch, H.H. The medical challenge to infrared spectroscopy. J. Mol. Struct. 408, 105–111 (1997).
Naumann, D., Helm, D. & Labischinski, H. Microbiological characterizations by FT-IR spectroscopy. Nature 351, 81–82 (1991).
Wong, P.T.T., Wong, R.K., Caputo, T.A., Godwin, T.A. & Rigas, B. Infrared spectroscopy of exfoliated human cervical cells—evidence of extensive structural changes during carcinogenesis. Proc. Natl. Acad. Sci. USA 88, 10988–10992 (1991).
Malins, D.C., Polissar, N.L. & Gunselman, S.J. Models of DNA structure achieve almost perfect discrimination between normal prostate, benign prostatic hyperplasia (BPH), and adenocarcinoma and have a high potential for predicting BPH and prostate cancer. Proc. Natl. Acad. Sci. USA 94, 259–264 (1997).
Andrus, P.G.L. & Strickland, R.D. Cancer grading by Fourier transform infrared spectroscopy. Biospectroscopy 4, 37–46 (1998).
Heise, H.M., Kupper, L. & Butvina, L.N. Bio-analytical applications of mid-infrared spectroscopy using silver halide fiber-optic probes. Spectrochim. Acta B 57, 1649–1663 (2002).
Bhargava, R. & Levin, I.W. Fourier transform infrared imaging: theory and practice. Anal. Chem. 73, 5157–5167 (2001).
Ou-Yang, H., Paschalis, E.P., Mayo, W.E., Boskey, A.L. & Mendelsohn, R. Infrared microscopic imaging of bone: spatial distribution of CO32−. J. Bone Miner. Res. 16, 893–900 (2001).
Kidder, L.H., Kalasinsky, V.F., Luke, J.L., Levin, I.W. & Lewis, E.N. Visualization of silicone gel in human breast tissue using new infrared imaging spectroscopy. Nat. Med. 3, 235–237 (1997).
Levin, I.W. & Bhargava, R. Fourier transform infrared vibrational spectroscopic imaging: integrating microscopy and molecular recognition. Annu. Rev. Phys. Chem. 56, 429–474 (2005).
Wood, B.R. et al. Fourier transform infrared (FTIR) spectral mapping of the cervical transformation zone, and dysplastic squamous epithelium. Gynecol. Oncol. 93, 59–68 (2004).
Malins, D.C. et al. The etiology and prediction of breast-cancer—Fourier transform-infrared spectroscopy reveals progressive alterations in breast DNA leading to a cancer-like phenotype in a high proportion of normal women. Cancer 75, 503–517 (1995).
Boydston-White, S., Gopen, T., Houser, S., Bargonetti, J. & Diem, M. Infrared spectroscopy of human tissue. V. Infrared spectroscopic studies of myeloid leukemia (ML-1) cells at different phases of the cell cycle. Biospectroscopy 5, 219–227 (1999).
Shaw, R.A., Guijon, F.B., Paraskevas, M., Ying, S.L. & Mantsch, H.H. Infrared spectroscopy of exfoliated cervical cell specimens—proceed with caution. Anal. Quant. Cytol. 21, 292–302 (1999).
Mansfield, J.R., McIntosh, L.M., Crowson, A.N., Mantsch, H.H. & Jackson, M. LDA-guided search engine for the nonsubjective analysis of infrared microscopic maps. Appl. Spectrosc. 53, 1323–1333 (1999).
McIntosh, L.M. et al. Infrared spectra of basal cell carcinomas are distinct from non-tumor-bearing skin components. J. Invest. Dermatol. 112, 951–956 (1999).
Webb, A.R. Statistical Pattern Recognition (John Wiley & Sons, New York, 2002).
Goodacre, R. Explanatory analysis of spectroscopic data using machine learning of simple, interpretable rules. Vib. Spectrosc. 32, 33–45 (2003).
Lasch, P. et al. Characterization of colorectal adenocarcinoma sections by spatially resolved FT-IR microspectroscopy. Appl. Spectrosc. 56, 1–9 (2002).
Kwiatkoski, J.M. & Reffner, J.A. FT-IR microspectrometry advances. Nature 328, 837–838 (1987).
Wetzel, D.L. & LeVine, S.M. Microspectroscopy—imaging molecular chemistry with infrared microscopy. Science 285, 1224–1225 (1999).
Liotta, L.A. & Kohn, E.C. The microenvironment of the tumour-host interface. Nature 411, 375–379 (2001).
Tuxhorn, J.A. et al. Reactive stroma in human prostate cancer: induction of myofibroblast phenotype and extracellular matrix remodeling. Clin. Cancer Res. 8, 2912–2923 (2002).
Swets, J.A. Measuring the accuracy of diagnostic systems. Science 240, 1285–1293 (1988).
Bradley, A.P. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recogn. 30, 1145–1159 (1997).
Rubin, M.A. Use of laser capture microdissection, cDNA microarrays, and tissue microarrays in advancing our understanding of prostate cancer. J. Pathol. 195, 80–86 (2001).
Acknowledgements
We thank Kimberly Tuttle for technical assistance in preparing tissue microarrays and samples for spectroscopic imaging.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
H&E stained images and their corresponding classified images from two arrays demonstrating the validity of the approach for large populations. (PDF 740 kb)
Supplementary Fig. 2
Patient-matched benign and malignant samples visualized using conventional staining and spectroscopic segmentation. (PDF 245 kb)
Supplementary Table 1
Spectral biomarkers employed for spectroscopic histology. (PDF 10 kb)
Supplementary Table 2
Spectral biomarkers employed for distinguishing diseased from benign epithelial tissue. (PDF 12 kb)
Rights and permissions
About this article
Cite this article
Fernandez, D., Bhargava, R., Hewitt, S. et al. Infrared spectroscopic imaging for histopathologic recognition. Nat Biotechnol 23, 469–474 (2005). https://doi.org/10.1038/nbt1080
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1080
This article is cited by
-
Infrared spectroscopic laser scanning confocal microscopy for whole-slide chemical imaging
Nature Communications (2023)
-
Towards a point-of-care multimodal spectroscopy instrument for the evaluation of human cardiac tissue
Heart and Vessels (2023)
-
Effects of the coupling of dielectric spherical particles on signatures in infrared microspectroscopy
Scientific Reports (2022)
-
High-throughput analysis of tissue microarrays using automated desorption electrospray ionization mass spectrometry
Scientific Reports (2022)
-
Recent Advances in Sensor-Based Detection of Toxic Dyes for Bioremediation Application: a Review
Applied Biochemistry and Biotechnology (2022)