Abstract
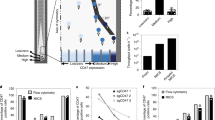
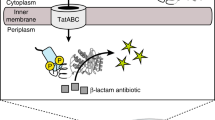
Here we describe a high-throughput, quantitative method for the isolation of enzymes with novel substrate specificities from large libraries of protein variants. Protein variants are displayed on the surface of microorganisms and incubated with a synthetic substrate consisting of (1) a fluorescent dye (2) a positively charged moiety (3) the target scissile bond, and (4) a fluorescence resonance energy transfer (FRET) quenching partner. Enzymatic cleavage of the scissile bond results in release of the FRET quenching partner while the fluorescent product is retained on the cell surface, allowing isolation of catalytically active clones by fluorescence-activated cell sorting (FACS). Using a synthetic substrate with these characteristics, we enriched Escherichia coli expressing the serine protease OmpT from cells expressing an inactive OmpT variant by over 5,000-fold in a single round. Screening a library of 6 × 105 random OmpT variants by FACS using a FRET peptide substrate with a nonpreferred Arg-Val cleavage sequence resulted in the isolation of variant proteases with catalytic activities enhanced by as much as 60-fold. This approach represents a potentially widely applicable method for high-throughput screening of large libraries on the basis of catalytic turnover.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kast, P. & Hilvert, D. 3D structural information as a guide to protein engineering using genetic selection. Curr. Opin. Struct. Biol. 7, 470–479 (1997).
Arnold, F.H. & Volkov, A.A. Directed evolution of biocatalysts. Curr. Opin. Chem. Biol. 3, 54–59 (1999).
Stemmer, W.P.C. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Nature 370, 389–391 (1994).
Crameri, A., Raillard, S.A., Bermudez, E. & Stemmer, W.P.C. DNA shuffling of a family of genes from diverse species accelerates directed evolution. Nature 391, 288–291 (1998).
Joo, H., Lin, Z. & Arnold, F.H. Laboratory evolution of peroxide-mediated cytochrome P450 hydroxylation. Nature 399, 670–673 (1998).
Matsumura, I., Wallingford, J.B., Surana, N.K., Vize, P.D. & Ellington, A.D. Directed evolution of the surface chemistry of the reporter enzyme beta-glucuronidase. Nat. Biotechnol. 17, 696–701 (1999).
MacBeath, G., Kast, P. & Hilvert, D. Redesigning enzyme topology by directed evolution. Science 279, 1958–1961 (1998).
Cantu, C., Huang, W. & Palzkill, T, Cephalosporin substrate specificity determinants of TEM-1 beta-lactamase. J. Biol. Chem. 272, 29144–29150 (1997).
Smiley, J.A. & Benkovic, S.J. Selection of catalytic antibodies for a biosynthetic reaction from a combinatorial cDNA library by complementation of an auxotrophic Escherichia coli: antibodies for orotate decarboxylation. Proc. Natl. Acad. Sci. USA 91, 8319–8323 (1994).
Christians, F.C. & Loeb, L.A. Novel human DNA alkyltransferases obtained by random substitution and genetic selection in bacteria. Proc. Natl. Acad. Sci. USA 93, 6124–6128 (1996).
Yano, T., Oue, S. & Kagamiyama, H. Directed evolution of an aspartate aminotransferase with new substrate specificities Proc. Natl. Acad. Sci. USA 95, 5511–5515 (1998).
Rader, C. & Barbas, C.F. Phage display of combinatorial antibody libraries. Curr. Opin. Biotechnol. 8, 503–508 (1997).
Demartis, S. et al. A strategy for the isolation of catalytic activities from repertoires of enzymes displayed on phage. J. Mol. Biol. 286, 617–633 (1999).
Pedersen, H. et al. A method for directed evolution and functional cloning of enzymes. Proc. Natl. Acad. Sci. USA 95, 10523–1058 (1998).
Atwell, S. & Wells, J.A. Selection for improved subtiligases by phage display. Proc. Natl. Acad. Sci. USA 96, 9497–9502 (1999).
Georgiou, G. et al. Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nat. Biotechnol. 15, 29–34 (1997).
Razatos, A., Ong, Y.L., Sharma, M.M. & Georgiou, G. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Nat. Acad. Sci. USA 95, 11059–11064 (1998).
Daugherty, P.S., Olsen, M.J., Iverson, B.L. & Georgiou, G. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. 12, 613–621 (1999).
Kamentsky, L.A., Melamed, M.R. & Derman, H. Spectrophotometer: new instrument for ultrarapid cell analysis Science 150, 630–631 (1965).
Hulett, H.R., Bonner, W.A., Barrett, J. & Herzenberg, L.A. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science 166, 747–749 (1969).
Shapiro, H.M. Practical flow cytometry, Edn. 3 (Wiley-Liss, New York, NY; 1995).
Leytus, S.P., Bowles, L.K., Konisky, J. & Mangel, W.F. Activation of plasminogen to plasmin by a protease associated with the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 78, 1485–1489 (1981).
Sodeinde, O.A. et al. A surface protease and the invasive character of plague. Science 25, 1004–1007 (1992).
Sugimura, K. & Nishihara, T. Purification, characterization, and primary structure of Escherichia coli protease VII with specificity for paired basic residues: identity of protease VII and OmpT. J. Bacteriol. 170, 5625–5632 (1988).
White, C.B., Chen, Q., Kenyon, G.L. & Babbitt, P.C. A novel activity of OmpT. Proteolysis under extreme denaturing conditions J. Biol. Chem. 270, 12990–12994 (1995).
Daugherty, P.S., Gang, C., Iverson, B.L. & Georgiou G. Quantitative analysis of the effect of the mutation frequency on the affinity maturation of single chain Fv antibodies. Proc. Natl Acad. Sci USA 97, 2029–2034 (2000).
Rashtchian, A., Buchman, G.W., Schuster, D.M. & Berninger, M.S. Uracil DNA glycosylase-mediated cloning of polymerase chain reaction-amplified DNA: application to genomic and cDNA cloning. Anal. Biochem. 206, 91–97 (1992).
Mangel, W.F. et al. Omptin: an Escherichia coli outer membrane proteinase that activates plasminogen. Methods Enzymol. 244, 384–399 (1994).
Grodberg, J., Lundrigan, M.D., Toledo, D.L., Mangel, W.F. & Dunn, J.J. Complete nucleotide sequence and deduced amino acid sequence of the ompT gene of Escherichia coli K-12. Nucleic Acids Res. 16, 1209 (1988).
Deng, W.P. & Nickoloff, J.A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem. 200, 81–88 (1992).
Cadwell, R.C. & Joyce, G.F. Randomization of genes by PCR mutagenesis. PCR Methods Appl. 2, 128–133 (1992).
Acknowledgements
We would like to thank W.F. Mangel, M. Ostermeier, and C.B. Mullins for reading the manuscript and for many useful comments. We also thank the University of Texas Peptide Synthesis Facility, and the University of Texas Mass Spectrometer Facility. This work was supported by grants from the National Science Foundation BES, DARPA(Defense Advanced Research Projects Agency)(MDA972-97-10009), US Department of Energy (DE-96ER62322), Texas Higher Education Coordinating Board (ATP 3658-0423-1999), and the Welch Foundation (G.G.).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Olsen, M., Stephens, D., Griffiths, D. et al. Function-based isolation of novel enzymes from a large library. Nat Biotechnol 18, 1071–1074 (2000). https://doi.org/10.1038/80267
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/80267
This article is cited by
-
A mixed culture of bacterial cells enables an economic DNA storage on a large scale
Communications Biology (2020)
-
Rapid micro-assays for amylolytic activities determination: customization and validation of the tests
Applied Microbiology and Biotechnology (2019)
-
Single-cell analysis and sorting using droplet-based microfluidics
Nature Protocols (2013)
-
Directed evolution of cutinase using in vitro compartmentalization
Biotechnology and Bioprocess Engineering (2012)
-
Development of an enzyme activity screening system for β-glucosidase-displaying yeasts using calcium alginate micro-beads and flow sorting
Applied Microbiology and Biotechnology (2009)