Abstract
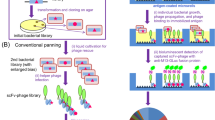
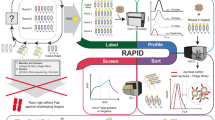
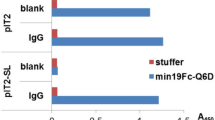
Periplasmic expression with cytometric screening (PECS) is a powerful and rapid “display-less” technology for isolating ligand-binding proteins from diverse libraries. Escherichia coli expressing a library of proteins secreted into the periplasmic space are incubated with a fluorescent conjugate of the target ligand. Under the proper conditions, ligands as large as about 10 kDa can equilibrate within the periplasmic space without compromising the cell's integrity or viability. The bacterial cell envelope effectively serves as a dialysis bag to selectively retain receptor–fluorescent probe complexes but not free ligand. Cells displaying increased fluorescence are then isolated by flow cytometry. We demonstrate that scFv antibodies with both very high and low affinity to digoxigenin can be isolated from libraries screened by PECS using a benchtop flow cytometer. We also show that preexisting libraries constructed for display on filamentous bacteriophage can be screened by PECS without the need for subcloning. In fact, PECS was found to select for proteins that could be missed by conventional phage panning and screening methods.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Shusta, E.V., VanAntwerp, J. & Wittrup, K.D. Biosynthetic polypeptide libraries. Curr. Opin. Biotechnol. 10, 117–122 (1999).
Rodi, D.J. & Makowski, L. Phage display technology—finding a needle in a vast molecular haystack. Curr. Opin. Biotechnol. 10, 87–93 (1999).
Boder, E.T. & Wittrup, K.D. Yeast surface display for directed evolution of protein expression, affinity, and stability. Methods Enzymol. 328, 430–444 (2000).
Daugherty, P.S., Iverson, B.L. & Georgiou, G. Flow cytometric screening of cell-based libraries. J. Immunol. Methods 243, 211–227 (2000).
Olsen, M.J. et al. Function-based isolation of novel enzymes from a large library. Nat. Biotechnol. 18, 1071–1074 (2000).
Maenaka, K. et al. A stable phage-display system using a phagemid vector: phage display of hen egg-white lysozyme (HEL), Escherichia coli alkaline, phosphatase, and anti-HEL monoclonal antibody, HyHEL10. Biochem. Biophys. Res. Commun. 218, 682–687 (1996).
Corey, D.R., Shiau, A.K., Yang, Q., Janowski, B.A. & Craik, C.S. Trypsin display on the surface of bacteriophage. Gene 128, 129–134 (1993).
Weiner, J.H. et al. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93, 93–101 (1998).
Deng, S.J. et al. Basis for selection of improved carbohydrate-binding single-chain antibodies from synthetic gene libraries. Proc. Natl. Acad. Sci. USA 92, 4992–4996 (1995).
MacKenzie, C.R. et al. Analysis by surface plasmon resonance of the influence of valence on the ligand binding affinity and kinetics of an anti-carbohydrate antibody. J. Biol. Chem. 271, 1527–1533 (1996).
MacKenzie, R. & To, R. The role of valency in the selection of anti-carbohydrate single-chain Fvs from phage display libraries. J. Immunol. Methods 220, 39–49 (1998).
Decad, G.M. & Nikaido, H. Outer membrane of gram-negative bacteria. XII. Molecular-sieving function of cell wall. J. Bacteriol. 128, 325–336 (1976).
Chen, G., Dubrawsky, I., Mendez, P., Georgiou, G. & Iverson, B.L. In vitro scanning saturation mutagenesis of all the specificity determining residues in an antibody binding site. Protein Eng. 12, 349–356 (1999).
Schildbach, J.F. et al. Heavy chain position 50 is a determinant of affinity and specificity for the anti-digoxin antibody 26-10. J. Biol. Chem. 268, 21739–21747 (1993).
Short, M.K., Jeffrey, P.D., Kwong, R.F. & Margolies, M.N. Contribution of antibody heavy chain CDR1 to digoxin binding analyzed by random mutagenesis of phage-displayed Fab 26-10. J. Biol. Chem. 270, 28541–28550 (1995).
Daugherty, P.S., Chen, G., Olsen, M.J., Iverson, B.L. & Georgiou, G. Antibody affinity maturation using bacterial surface display. Protein Eng. 11, 825–832 (1998).
Jeffrey, P.D. et al. 26-10 Fab–digoxin complex: affinity and specificity due to surface complementarity. Proc. Natl. Acad. Sci. USA 90, 10310–10314 (1993).
Boder, E.T. & Wittrup, K.D. Optimal screening of surface-displayed polypeptide libraries. Biotechnol. Prog. 14, 55–62 (1998).
Gough, K.C. et al. Selection of phage antibodies to surface epitopes of Phytophthora infestans. J. Immunol. Methods 228, 97–108 (1999).
Martinez, M.B., Flickinger, M.C. & Nelsestuen, G.L. Steady-state enzyme kinetics in the Escherichia coli periplasm: a model of a whole cell biocatalyst. J. Biotechnol. 71, 59–66 (1999).
Homma, T. & Nakae, T. Effects of cations on the outer membrane permeability of Escherichia coli. Tokai J. Exp. Clin. Med. 7, 171–175 (1982).
Vaara, M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 56, 395–411 (1992).
Boeke, J.D., Model, P. & Zinder, N.D. Effects of bacteriophage f1 gene III protein on the host cell membrane. Mol. Gen. Genet. 186, 185–192 (1982).
Krebber, A. et al. Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Methods. 201, 35–55 (1997).
Marciano, D.K., Russel, M. & Simon, S.M. An aqueous channel for filamentous phage export. Science 284, 1516–1519. (1999).
Brissette, J.L., Russel, M., Weiner, L. & Model, P. Phage shock protein, a stress protein of Escherichia coli. Proc. Natl. Acad. Sci. USA 87, 862–866. (1990).
Hayhurst, A. & Harris, W.J. Escherichia coli skp chaperone coexpression improves solubility and phage display of single-chain antibody fragments. Protein Expr. Purif. 15, 336–343 (1999).
Rakonjac, J. & Model, P. Roles of pIII in filamentous phage assembly. J. Mol. Biol. 282, 25–41 (1998).
Rakonjac, J., Feng, J. & Model, P. Filamentous phage are released from the bacterial membrane by a two- step mechanism involving a short C-terminal fragment of pIII. J. Mol. Biol. 289, 1253–1265 (1999).
Georgiou, G. Analysis of large libraries of protein mutants using flow cytometry. Adv. Protein Chem. 55, 293–315 (2000).
Fu, A.Y., Spence, C., Scherer, A., Arnold, F.H. & Quake, S.R. A microfabricated fluorescence-activated cell sorter. Nat. Biotechnol. 17, 1109–1111 (1999).
Daugherty, P.S., Olsen, M.J., Iverson, B.L. & Georgiou, G. Development of an optimized expression system for the screening of antibody libraries displayed on the Escherichia coli surface. Protein Eng. 12, 613–621 (1999).
Acknowledgements
We thank the Greg Winter laboratory, Medical Research Council, UK for the Griffin.1 library, Charles H. Patterson Jr. Center for Bio/Molecular Science and Engineering, Naval Research Laboratory (Washington, DC) for the Cy5-TNB probe, and the Jon Beckwith laboratory for strains. We thank Patrick S. Daugherty for critical reading of the manuscript and members of the Georgiou and Iverson laboratories for help and comment. This work was supported by Defense Advanced Research Projects Agency (DARPA). B.R.H. was supported by a US Army Multidisciplinary Research Program of the University Research Initiative (MURI) grant.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Chen, G., Hayhurst, A., Thomas, J. et al. Isolation of high-affinity ligand-binding proteins by periplasmic expression with cytometric screening (PECS). Nat Biotechnol 19, 537–542 (2001). https://doi.org/10.1038/89281
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/89281
This article is cited by
-
A novel Ffu fusion system for secretory expression of heterologous proteins in Escherichia coli
Microbial Cell Factories (2017)
-
A generic selection system for improved expression and thermostability of G protein-coupled receptors by directed evolution
Scientific Reports (2016)
-
Optimizing antibody expression by using the naturally occurring framework diversity in a live bacterial antibody display system
Scientific Reports (2015)
-
Auto-induction-based Rapid Evaluation of Extracellular Enzyme Expression from Lac Operator-involved Recombinant Escherichia coli
Applied Biochemistry and Biotechnology (2014)
-
Computational design of ligand-binding proteins with high affinity and selectivity
Nature (2013)