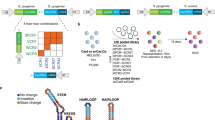
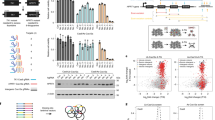
Abstract
Combinatorial genetic screening using CRISPR–Cas9 is a useful approach to uncover redundant genes and to explore complex gene networks. However, current methods suffer from interference between the single-guide RNAs (sgRNAs) and from limited gene targeting activity. To increase the efficiency of combinatorial screening, we employ orthogonal Cas9 enzymes from Staphylococcus aureus and Streptococcus pyogenes. We used machine learning to establish S. aureus Cas9 sgRNA design rules and paired S. aureus Cas9 with S. pyogenes Cas9 to achieve dual targeting in a high fraction of cells. We also developed a lentiviral vector and cloning strategy to generate high-complexity pooled dual-knockout libraries to identify synthetic lethal and buffering gene pairs across multiple cell types, including MAPK pathway genes and apoptotic genes. Our orthologous approach also enabled a screen combining gene knockouts with transcriptional activation, which revealed genetic interactions with TP53. The “Big Papi” (paired aureus and pyogenes for interactions) approach described here will be widely applicable for the study of combinatorial phenotypes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Roguev, A. et al. Quantitative genetic-interaction mapping in mammalian cells. Nat. Methods 10, 432–437 (2013).
Horn, T. et al. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat. Methods 8, 341–346 (2011).
Tong, A.H.Y. et al. Global mapping of the yeast genetic interaction network. Science 303, 808–813 (2004).
Costanzo, M. et al. A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 (2016).
Bassik, M.C. et al. A systematic mammalian genetic interaction map reveals pathways underlying ricin susceptibility. Cell 152, 909–922 (2013).
Wong, A.S.L. et al. Multiplexed barcoded CRISPR-Cas9 screening enabled by CombiGEM. Proc. Natl. Acad. Sci. USA 113, 2544–2549 (2016).
Shen, J.P. et al. Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat. Methods 14, 573–576 (2017).
Han, K. et al. Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol. 35, 463–474 (2017).
ter Brake, O. et al. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol. Ther. 16, 557–564 (2008).
Vidigal, J.A. & Ventura, A. Rapid and efficient one-step generation of paired gRNA CRISPR-Cas9 libraries. Nat. Commun. 6, 8083 (2015).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882.e21 (2016).
Zetsche, B. et al. Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat. Biotechnol. 35, 31–34 (2017).
McIntyre, G.J., Arndt, A.J., Gillespie, K.M., Mak, W.M. & Fanning, G.C. A comparison of multiple shRNA expression methods for combinatorial RNAi. Genet. Vaccines Ther. 9, 9 (2011).
Stockman, V.B. et al. A high-throughput strategy for dissecting mammalian genetic interactions. PLoS One 11, e0167617–e13 (2016).
Gilbert, L.A. et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661 (2014).
Doench, J.G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Zhu, S. et al. Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol. 34, 1279–1286 (2016).
Doench, J.G. et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 32, 1262–1267 (2014).
Ran, F.A. et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 520, 186–191 (2015).
Friedland, A.E. et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 16, 257 (2015).
Hanzlikova, H., Gittens, W., Krejcikova, K., Zeng, Z. & Caldecott, K.W. Overlapping roles for PARP1 and PARP2 in the recruitment of endogenous XRCC1 and PNKP into oxidized chromatin. Nucleic Acids Res. 45, 2546–2557 (2017).
Farmer, H. et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434, 917–921 (2005).
van Delft, M.F. et al. The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10, 389–399 (2006).
Sun, C. & Bernards, R. Feedback and redundancy in receptor tyrosine kinase signaling: relevance to cancer therapies. Trends Biochem. Sci. 39, 465–474 (2014).
Buscà, R., Pouysségur, J. & Lenormand, P. ERK1 and ERK2 Map Kinases: Specific Roles or Functional Redundancy? Front. Cell Dev. Biol. 4, 53 (2016).
Uzgare, A.R. & Isaacs, J.T. Enhanced redundancy in Akt and mitogen-activated protein kinase-induced survival of malignant versus normal prostate epithelial cells. Cancer Res. 64, 6190–6199 (2004).
Srivas, R. et al. A network of conserved synthetic lethal interactions for exploration of precision cancer therapy. Mol. Cell 63, 514–525 (2016).
Hart, T. et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526 (2015).
Chaudhuri, L. et al. CHK1 and WEE1 inhibition combine synergistically to enhance therapeutic efficacy in acute myeloid leukemia ex vivo. Haematologica 99, 688–696 (2014).
Souers, A.J. et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat. Med. 19, 202–208 (2013).
Oltersdorf, T. et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 (2005).
Leverson, J.D. et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci. Transl. Med. 7, 279ra40 (2015).
Lessene, G. et al. Structure-guided design of a selective BCL-X(L) inhibitor. Nat. Chem. Biol. 9, 390–397 (2013).
Kotschy, A. et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 538, 477–482 (2016).
Dummler, B. et al. Life with a single isoform of Akt: mice lacking Akt2 and Akt3 are viable but display impaired glucose homeostasis and growth deficiencies. Mol. Cell. Biol. 26, 8042–8051 (2006).
Nitulescu, G.M. et al. Akt inhibitors in cancer treatment: The long journey from drug discovery to clinical use (Review). Int. J. Oncol. 48, 869–885 (2016).
Stordal, B. et al. BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only one functionally deleterious BRCA1 mutation. Mol. Oncol. 7, 567–579 (2013).
Helleday, T. The underlying mechanism for the PARP and BRCA synthetic lethality: clearing up the misunderstandings. Mol. Oncol. 5, 387–393 (2011).
Rooswinkel, R.W. et al. Antiapoptotic potency of Bcl-2 proteins primarily relies on their stability, not binding selectivity. Blood 123, 2806–2815 (2014).
Cowling, V. & Downward, J. Caspase-6 is the direct activator of caspase-8 in the cytochrome c-induced apoptosis pathway: absolute requirement for removal of caspase-6 prodomain. Cell Death Differ. 9, 1046–1056 (2002).
Kroemer, G., Galluzzi, L. & Brenner, C. Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163 (2007).
Li, P. et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91, 479–489 (1997).
Ma, S.B. et al. Bax targets mitochondria by distinct mechanisms before or during apoptotic cell death: a requirement for VDAC2 or Bak for efficient Bax apoptotic function. Cell Death Differ. 21, 1925–1935 (2014).
Sarosiek, K.A. et al. BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol. Cell 51, 751–765 (2013).
Chavez, A. et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 12, 326–328 (2015).
Rotem, A. et al. Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc. Natl. Acad. Sci. USA 112, 5708–5713 (2015).
Miura, Y. et al. Susceptibility to killer T cells of gastric cancer cells enhanced by Mitomycin-C involves induction of ATBF1 and activation of p21 (Waf1/Cip1) promoter. Microbiol. Immunol. 48, 137–145 (2004).
Wong, C.C. et al. Inactivating CUX1 mutations promote tumorigenesis. Nat. Genet. 46, 33–38 (2014).
Faraonio, R. et al. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J. Biol. Chem. 281, 39776–39784 (2006).
Gao, Y. et al. Complex transcriptional modulation with orthogonal and inducible dCas9 regulators. Nat. Methods 13, 1043–1049 (2016).
Kleinstiver, B.P. et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 33, 1293–1298 (2015).
Lawrence, M.S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501 (2014).
Forbes, S.A. et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res. 45, D1, D777–D783 (2017).
Acknowledgements
We thank S. Elmore and G. Wei for helpful discussions; E. Sukharevsky, R. Hanna, and I. Sebenius for experimental assistance and comments on the manuscript; D. Ortiz for emotional uplift. We thank T. Hart and other anonymous referees for helpful comments during the review process. F.J.N. is supported by a cellular and developmental biology training grant (NIH T32GM007226-41). J.G.D. is supported by the Next Generation Fund at the Broad Institute of MIT and Harvard. This work was supported by the Functional Genomics Consortium (D.E.R.)., Starr Cancer Consortium (B.E.B.), and National Cancer Institute–NIH Common Fund (DP1CA216873) (B.E.B.).
Author information
Authors and Affiliations
Contributions
F.J.N., C.S., K.F.D., K.R.S., Z.K., E.W.V., M.E.S., E.H., and J.G.D. designed and performed experiments. N.F. and J.L. performed the computational modeling of SaCas9 activity. M.H., S.T.Y., and J.G.D. analyzed screening data. B.E.B. and D.E.R. provided senior guidance. F.J.N., D.E.R., and J.G.D. wrote the manuscript with assistance from other authors.
Corresponding author
Ethics declarations
Competing interests
J.L. and N.F. are employed by Microsoft Research. J.G.D. consults for Tango Therapeutics.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–12, Supplementary Note 1 and Supplementary Discussion (PDF 11619 kb)
Rights and permissions
About this article
Cite this article
Najm, F., Strand, C., Donovan, K. et al. Orthologous CRISPR–Cas9 enzymes for combinatorial genetic screens. Nat Biotechnol 36, 179–189 (2018). https://doi.org/10.1038/nbt.4048
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.4048
This article is cited by
-
A dual sgRNA library design to probe genetic modifiers using genome-wide CRISPRi screens
BMC Genomics (2023)
-
Optimized metrics for orthogonal combinatorial CRISPR screens
Scientific Reports (2023)
-
Massively parallel evaluation and computational prediction of the activities and specificities of 17 small Cas9s
Nature Methods (2023)
-
Revolutionizing DNA repair research and cancer therapy with CRISPR–Cas screens
Nature Reviews Molecular Cell Biology (2023)
-
Chromatin complex dependencies reveal targeting opportunities in leukemia
Nature Communications (2023)