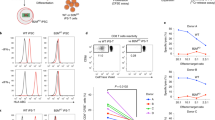
Abstract
Polymorphisms in the human leukocyte antigen (HLA) class I genes can cause the rejection of pluripotent stem cell (PSC)-derived products in allogeneic recipients. Disruption of the Beta-2 Microglobulin (B2M) gene eliminates surface expression of all class I molecules, but leaves the cells vulnerable to lysis by natural killer (NK) cells. Here we show that this 'missing-self' response can be prevented by forced expression of minimally polymorphic HLA-E molecules. We use adeno-associated virus (AAV)-mediated gene editing to knock in HLA-E genes at the B2M locus in human PSCs in a manner that confers inducible, regulated, surface expression of HLA-E single-chain dimers (fused to B2M) or trimers (fused to B2M and a peptide antigen), without surface expression of HLA-A, B or C. These HLA-engineered PSCs and their differentiated derivatives are not recognized as allogeneic by CD8+ T cells, do not bind anti-HLA antibodies and are resistant to NK-mediated lysis. Our approach provides a potential source of universal donor cells for applications where the differentiated derivatives lack HLA class II expression.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Taylor, C.J., Peacock, S., Chaudhry, A.N., Bradley, J.A. & Bolton, E.M. Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient HLA types. Cell Stem Cell 11, 147–152 (2012).
Nishikawa, S., Goldstein, R.A. & Nierras, C.R. The promise of human induced pluripotent stem cells for research and therapy. Nat. Rev. Mol. Cell Biol. 9, 725–729 (2008).
Taylor, C.J. et al. Banking on human embryonic stem cells: estimating the number of donor cell lines needed for HLA matching. Lancet 366, 2019–2025 (2005).
Braciale, T.J. Antigen processing for presentation by MHC class I molecules. Curr. Opin. Immunol. 4, 59–62 (1992).
Arce-Gomez, B., Jones, E.A., Barnstable, C.J., Solomon, E. & Bodmer, W.F. The genetic control of HLA-A and B antigens in somatic cell hybrids: requirement for beta2 microglobulin. Tissue Antigens 11, 96–112 (1978).
Riolobos, L. et al. HLA engineering of human pluripotent stem cells. Mol. Ther. 21, 1232–1241 (2013).
Lu, P. et al. Generating hypoimmunogenic human embryonic stem cells by the disruption of beta 2-microglobulin. Stem Cell Rev. 9, 806–813 (2013).
Wang, D., Quan, Y., Yan, Q., Morales, J.E. & Wetsel, R.A. Targeted disruption of the β2-microglobulin gene minimizes the immunogenicity of human embryonic stem cells. Stem Cells Transl. Med. 4, 1234–1245 (2015).
Feng, Q. et al. Scalable generation of universal platelets from human induced pluripotent stem cells. Stem Cell Reports 3, 817–831 (2014).
Bix, M. et al. Rejection of class I MHC-deficient haemopoietic cells by irradiated MHC-matched mice. Nature 349, 329–331 (1991).
Liao, N.S., Bix, M., Zijlstra, M., Jaenisch, R. & Raulet, D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science 253, 199–202 (1991).
Zarcone, D., Tilden, A.B., Friedman, H.M. & Grossi, C.E. Human leukemia-derived cell lines and clones as models for mechanistic analysis of natural killer cell-mediated cytotoxicity. Cancer Res. 47, 2674–2682 (1987).
Pazmany, L. et al. Protection from natural killer cell-mediated lysis by HLA-G expression on target cells. Science 274, 792–795 (1996).
Braud, V.M. et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998).
Lee, N. et al. HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. USA 95, 5199–5204 (1998).
Lilienfeld, B.G., Crew, M.D., Forte, P., Baumann, B.C. & Seebach, J.D. Transgenic expression of HLA-E single chain trimer protects porcine endothelial cells against human natural killer cell-mediated cytotoxicity. Xenotransplantation 14, 126–134 (2007).
Miller, J.D. et al. Analysis of HLA-E peptide-binding specificity and contact residues in bound peptide required for recognition by CD94/NKG2. J. Immunol. 171, 1369–1375 (2003).
Horowitz, A. et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med. 5, 208ra145 (2013).
Russell, D.W. & Hirata, R.K. Human gene targeting by viral vectors. Nat. Genet. 18, 325–330 (1998).
Khan, I.F., Hirata, R.K. & Russell, D.W. AAV-mediated gene targeting methods for human cells. Nat. Protoc. 6, 482–501 (2011).
Chamberlain, J.R. et al. Gene targeting in stem cells from individuals with osteogenesis imperfecta. Science 303, 1198–1201 (2004).
Li, L.B. et al. Trisomy correction in Down syndrome induced pluripotent stem cells. Cell Stem Cell 11, 615–619 (2012).
Khan, I.F. et al. Engineering of human pluripotent stem cells by AAV-mediated gene targeting. Mol. Ther. 18, 1192–1199 (2010).
Ichiryu, N. & Fairchild, P.J. Immune privilege of stem cells. Methods Mol. Biol. 1029, 1–16 (2013).
Pegram, H.J., Andrews, D.M., Smyth, M.J., Darcy, P.K. & Kershaw, M.H. Activating and inhibitory receptors of natural killer cells. Immunol. Cell Biol. 89, 216–224 (2011).
Gong, J.H., Maki, G. & Klingemann, H.G. Characterization of a human cell line (NK-92) with phenotypical and functional characteristics of activated natural killer cells. Leukemia 8, 652–658 (1994).
Brooks, A.G., Posch, P.E., Scorzelli, C.J., Borrego, F. & Coligan, J.E. NKG2A complexed with CD94 defines a novel inhibitory natural killer cell receptor. J. Exp. Med. 185, 795–800 (1997).
Brodin, P., Kärre, K. & Höglund, P. NK cell education: not an on-off switch but a tunable rheostat. Trends Immunol. 30, 143–149 (2009).
Huang, Y. et al. NK cells play a critical role in the regulation of class I-deficient hemopoietic stem cell engraftment: evidence for NK tolerance correlates with receptor editing. J. Immunol. 175, 3753–3761 (2005).
Rouas-Freiss, N., Gonçalves, R.M., Menier, C., Dausset, J. & Carosella, E.D. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc. Natl. Acad. Sci. USA 94, 11520–11525 (1997).
Zappacosta, F., Borrego, F., Brooks, A.G., Parker, K.C. & Coligan, J.E. Peptides isolated from HLA-Cw*0304 confer different degrees of protection from natural killer cell-mediated lysis. Proc. Natl. Acad. Sci. USA 94, 6313–6318 (1997).
Koller, B.H., Marrack, P., Kappler, J.W. & Smithies, O. Normal development of mice deficient in beta 2M, MHC class I proteins, and CD8+ T cells. Science 248, 1227–1230 (1990).
Zijlstra, M. et al. Beta 2-microglobulin deficient mice lack CD4-8+ cytolytic T cells. Nature 344, 742–746 (1990).
Mandal, P.K. et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell 15, 643–652 (2014).
DeSandro, A., Nagarajan, U.M. & Boss, J.M. The bare lymphocyte syndrome: molecular clues to the transcriptional regulation of major histocompatibility complex class II genes. Am. J. Hum. Genet. 65, 279–286 (1999).
Street, S.E.A. et al. Innate immune surveillance of spontaneous B cell lymphomas by natural killer cells and gammadelta T cells. J. Exp. Med. 199, 879–884 (2004).
Gadola, S.D., Moins-Teisserenc, H.T., Trowsdale, J., Gross, W.L. & Cerundolo, V. TAP deficiency syndrome. Clin. Exp. Immunol. 121, 173–178 (2000).
Pietra, G. et al. HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc. Natl. Acad. Sci. USA 100, 10896–10901 (2003).
Hansen, S.G. et al. Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351, 714–720 (2016).
Pietra, G., Romagnani, C., Manzini, C., Moretta, L. & Mingari, M.C. The emerging role of HLA-E-restricted CD8+ T lymphocytes in the adaptive immune response to pathogens and tumors. J. Biomed. Biotechnol. 2010, 907092 (2010).
Eichelberger, M., Allan, W., Zijlstra, M., Jaenisch, R. & Doherty, P.C. Clearance of influenza virus respiratory infection in mice lacking class I major histocompatibility complex-restricted CD8+ T cells. J. Exp. Med. 174, 875–880 (1991).
Hou, S., Doherty, P.C., Zijlstra, M., Jaenisch, R. & Katz, J.M. Delayed clearance of Sendai virus in mice lacking class I MHC-restricted CD8+ T cells. J. Immunol. 149, 1319–1325 (1992).
Spriggs, M.K. et al. Beta 2-microglobulin-, CD8+ T-cell-deficient mice survive inoculation with high doses of vaccinia virus and exhibit altered IgG responses. Proc. Natl. Acad. Sci. USA 89, 6070–6074 (1992).
Ware, C.B. et al. Derivation of naive human embryonic stem cells. Proc. Natl. Acad. Sci. USA 111, 4484–4489 (2014).
Thomson, J.A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Kotini, A.G. et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat. Biotechnol. 33, 646–655 (2015).
Leach, L.L., Buchholz, D.E., Nadar, V.P., Lowenstein, S.E. & Clegg, D.O. Canonical/β-catenin Wnt pathway activation improves retinal pigmented epithelium derivation from human embryonic stem cells. Invest. Ophthalmol. Vis. Sci. 56, 1002–1013 (2015).
Sambrook, J. & Russell, D.W. Molecular Cloning: a Laboratory Manual 3rd edn. (Cold Spring Harbor Laboratory Press, 2001).
Papapetrou, E.P. & Sadelain, M. Generation of transgene-free human induced pluripotent stem cells with an excisable single polycistronic vector. Nat. Protoc. 6, 1251–1273 (2011).
Lauterbach, N., Voorter, C.E.M. & Tilanus, M.G.J. Molecular typing of HLA-E. Methods Mol. Biol. 882, 143–158 (2012).
Aicheler, R.J. & Stanton, R.J. Functional NK cell cytotoxicity assays against virus infected cells. Methods Mol. Biol. 1064, 275–287 (2013).
Terasaki, P.I. & McClelland, J.D. Mirodroplet assay of human serum cytotoxins. Nature 204, 998–1000 (1964).
Vernon, R.B. et al. Reversal of diabetes in mice with a bioengineered islet implant incorporating a type I collagen hydrogel and sustained release of vascular endothelial growth factor. Cell Transplant. 21, 2099–2110 (2012).
Acknowledgements
This work was supported by US National Institutes of Health (NIH) grants DK55759 and HL007093 to D.W.R. 1S10OD010652-01 to the UW Shared IVIS Core Facility, and HL007093 to A.G.C. as well as California Institute for Regenerative Medicine grants DR3-07438, TG2-01161, TG2-01151, CL1-00521 and FA1-00616 to D.O.C. We also acknowledge support from the Garland Initiative for Vision, The Foundation Fighting Blindness Wynn-Gund Translational Research Acceleration Program and the UCSB Institute for Collaborative Biotechnologies through grant W911NF-09-0001 from the US Army Research Office. The authors thank R. Stolitenko for technical assistance, D. Geraghty (FHCRC) for providing HLA-E plasmids, D. McDonald (FHCRC) for cytogenetic analyses, D. Youngs (Bloodworks Northwest) for help in performing CDC assays, P. Treuting, B. Johnson and Y.-T. Tien (University of Washington) for histology and immunohistochemistry analysis, W. Loomis for assistance in IVIS image analysis, S. Quaratella (Medtronic Inc.) for providing PVA sponges, M. Gerace and D. Yadock (FHCRC) for providing CD8+ T and CD56+ NK cells (funding provided by the Cooperative Centers of Excellence in Hematology: NIH Grant DK106829)) and C. Levy (FHCRC) for help with graphing programs.
Author information
Authors and Affiliations
Contributions
G.G.G. and D.W.R. wrote the manuscript. G.G.G. and D.W.R. designed the experiments. G.G.G. and R.K.H. prepared AAV vectors stocks and performed gene editing. G.G.G. performed flow cytometry, hematopoietic differentiation, chromium release assays, CDC assays and data analysis. S.E.F. conducted in vivo experiments, PCR analysis and Southern blots. D.P. and R.K.H. performed flow sorting experiments. G.G.G., S.E.F. and G.M. did molecular cloning. G.M. performed RNA analysis. L.R. designed both the HLA-E dimer and trimer open reading frames, conducted preliminary studies with HLA-E vectors and optimized chromium release assay protocols. V.S.L., D.O.C. and G.G.G. performed RPE differentiation experiments and immunocytochemistry. L.-A.H. and C.T. did flow cytometry to detect costimulatory receptors. A.G.C. and L.R. each designed and built one of the AAV vectors. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
D.W.R. is on the Advisory Board of Horizon Discovery and co-founder of Universal Cells, Inc. D.O.C. is co-founder of Regenerative Patch Technologies, LLC.
Integrated supplementary information
Supplementary Figure 1 Generation of B2M-/- ESCs.
(a) Maps of the B2M alleles in cells with the indicated B2M genotypes. Probes and restriction enzymes used in Southern blots are indicated (H, Hind III; B, Bgl II; G, BsrG I). Dark blue regions show overlap with the gene editing vectors used to produce each allele. (b) B2M flow cytometry of different mixed colonies of ESCs and their subcloning results showing the bias against obtaining pure populations of normal B2M-/- cells. (c) Flow cytometry showing a lack of B2M expression in two B2M-/- ESC clones after IFN-γ treatment. (d) Karyotypic analysis of these same two B2M-/- clones.
Supplementary Figure 2 Trace B2M expression in B2M-edited clones.
(a) Pan-HLA- ABC flow cytometry of IFN-γ-treated B2M+/+ and B2M-/- H1 ESCs created with vectors AAV-B2M-EHyTKpA and AAV-B2M-ETKNpA6 (blue tracings) and isotype controls (red tracings) showing low level expression. (b) The sequence of an mRNA isolated from B2M-/- H1 ESCs showing transcription of the edited allele through the pA site, the location of an initiation codon (underlined) in the loxP site, and in-frame continuation of the reading frame into downstream B2M exon 1 sequences. (c) Modifications made in vectors AAV- B2M-HyTK and AAV-B2M-TKN that eliminate trace B2M expression. The B2M gene is shown after Cre-mediated excision of the HyTK or TKN genes present in either vector, with the loxP-encoded ATG start codon shown above, and the downstream stop codons that prevent translation (asterisks) in all three reading frames shown below.
Supplementary Figure 3 Retinal pigmented epithelium (RPE) cell differentiation.
RPE cells derived from B2M+/+ H9 ESCs (panels a-c) and B2M-/Edimer ESCs (panels d-f) were visualized by bright field microscopy (a, d) and immunofluorescence microscopy for retinal markers PMEL (b, e) and MITF (c, f) after 42 days of differentiation. Bright field images demonstrate the level of pigmentation. PMEL+ and MITF+ cells are shown in green, with DAPI stained nuclei in blue. Scale bar = 50 μm.
Supplementary Figure 4 Hematopoietic potential and NK cell-mediated lysis of ESC- derived CD45+ cells.
(a) Flow cytometry analysis of CD45 expression after hematopoietic differentiation of Elf-1 ESCs with the indicated B2M genotypes. Data were acquired from suspension cells on day 38 of differentiation. Results for B2M-/Etrimer c5 shown in Fig. 3B. (b) B2M-/- ESCs produce fewer hematopoietic cells. Kinetics of suspension cell production during hematopoietic differentiation of ESCs with the indicated B2M genotypes. Y-axis denotes number of live suspension cells generated per 5x106 undifferentiated ESCs. The results from two independent differentiation experiments are shown with numbers between parentheses. (c) Flow cytometry analysis of NKG2A and NKG2C receptors on NK cells derived from donor 2. Percents were calculated by subtracting the isotype control frequencies. (d) Chromium release assay with NK cells from donor 2 and ESC-derived CD45+ cells with the indicated B2M genotypes showing B2M-/Edimer expression partially prevents lysis by NK cells with low NKG2A expression levels. Data are represented as mean + SD (n=3). (e) Chromium release assay with NK cells from donor 1 and ESC- derived CD45+ cells showing that B2M-/- and B2M-/Edimer(pre-Cre) cells had similar susceptibility to NK-mediated lysis. Data are represented as mean + SD (n=3). (f) Chromium release assay as in (d) but with NK cells from donor 3 cultured at a low IL-2 dose (100 U/ml). Asterisks indicate p<0.05 for pair-wise comparison between the indicated cells (ANOVA followed by the post hoc Tukey HSD test). (g) Change in luciferase expression in B2M-/Edimer(pre-Cre) (HLA class I-negative control) and B2M-/Etrimer teratomas measured from day 13 to day 19 after implantation, with NK-92 cells administered to half the animals on days 13, 15 and 17. P-values were determined in each group (with or without NK-92 cells) by paired Student’s t-test (n=3 mice for both groups that received NK-92 cells and n=4 for both groups that did not). An unpaired Student’s t-test (p=0.096) was also applied to the same data to compare the relative growth of the B2M-/Etrimer and B2M-/Edimer(pre-Cre) teratomas in mice that received NK-92 cells to their relative growth in mice that did not receive NK-92 cells. (h) Examples of luciferase imaging in mice from (g), half of which received NK-92 cells as noted. “Pre” indicates B2M-/Edimer(pre-Cre) genotype, “-/E” indicates B2M-/Etrimer genotype. Red circles indicate measured areas.
Supplementary Figure 5 HLA molecule and costimulatory receptor expression.
(a) Flow cytometry analysis of HLA-ABC and HLA-DR expression in IFN-γ-stimulated B2M+/+ Elf-1 EBs used for priming CD8+ T cells as shown in Figure 4A. (b) Costimulatory receptor profile of B2M+/+ Elf-1 EBs. Isotype controls in red and specific antibodies in blue. (c) Costimulatory receptor profile for ESC-derived CD45+ cells with the indicated B2M genotypes.
Supplementary Figure 6 Differential growth of B2M+/+ and B2M-/Etrimer ESC-derived teratomas when challenged with allogeneic CD8+ T cells in vivo.
(a) Luciferase signal measured on day 1 was used to normalize the data. Each graph shows the results from an individual mouse. In all panels, blue and red lines show the growth of B2M-/Etrimer and B2M +/+ teratomas respectively. The three bottom panels show teratoma growth in mice that did not receive CD8+ cells. (b) B2M-/Etrimer and B2M+/+ teratoma growth rates (luciferase measurement increase per day) were measured before and after allogeneic primed CD8+ T cell infusions from three different donors (n=9 teratomas per genotype). Horizontal black bars indicate the means. P values were calculated by paired Student’s t-tests. (c) Quantitative measurement of CD8+ T cells in B2M-/Etrimer and B2M+/+ teratomas (n=5 mice, each with both types of teratoma; each line represents one mouse). Results were calculated as % of CD8 signal in the total area examined. The p value was calculated by paired Student’s t-test.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 and Supplementary Tables 1 and 2. (PDF 1663 kb)
Rights and permissions
About this article
Cite this article
Gornalusse, G., Hirata, R., Funk, S. et al. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat Biotechnol 35, 765–772 (2017). https://doi.org/10.1038/nbt.3860
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3860
This article is cited by
-
Hypoimmune cells resist rejection in monkeys
Nature Biotechnology (2024)
-
Allogeneic CAR-T cells with of HLA-A/B and TRAC disruption exhibit promising antitumor capacity against B cell malignancies
Cancer Immunology, Immunotherapy (2024)
-
An angiogenesis-associated gene-based signature predicting prognosis and immunotherapy efficacy of head and neck squamous cell carcinoma patients
Journal of Cancer Research and Clinical Oncology (2024)
-
Hypoimmune induced pluripotent stem cells survive long term in fully immunocompetent, allogeneic rhesus macaques
Nature Biotechnology (2024)
-
A highly efficient transgene knock-in technology in clinically relevant cell types
Nature Biotechnology (2024)