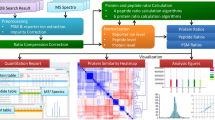
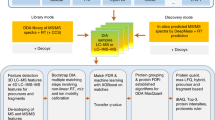
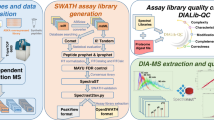
Abstract
Consistent and accurate quantification of proteins by mass spectrometry (MS)-based proteomics depends on the performance of instruments, acquisition methods and data analysis software. In collaboration with the software developers, we evaluated OpenSWATH, SWATH 2.0, Skyline, Spectronaut and DIA-Umpire, five of the most widely used software methods for processing data from sequential window acquisition of all theoretical fragment-ion spectra (SWATH)-MS, which uses data-independent acquisition (DIA) for label-free protein quantification. We analyzed high-complexity test data sets from hybrid proteome samples of defined quantitative composition acquired on two different MS instruments using different SWATH isolation-window setups. For consistent evaluation, we developed LFQbench, an R package, to calculate metrics of precision and accuracy in label-free quantitative MS and report the identification performance, robustness and specificity of each software tool. Our reference data sets enabled developers to improve their software tools. After optimization, all tools provided highly convergent identification and reliable quantification performance, underscoring their robustness for label-free quantitative proteomics.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Mallick, P. & Kuster, B. Proteomics: a pragmatic perspective. Nat. Biotechnol. 28, 695–709 (2010).
Distler, U., Kuharev, J. & Tenzer, S. Biomedical applications of ion mobility-enhanced data-independent acquisition-based label-free quantitative proteomics. Expert Rev. Proteomics 11, 675–684 (2014).
Gillet, L.C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012).
Geromanos, S.J., Hughes, C., Ciavarini, S., Vissers, J.P.C. & Langridge, J.I. Using ion purity scores for enhancing quantitative accuracy and precision in complex proteomics samples. Anal. Bioanal. Chem. 404, 1127–1139 (2012).
Geiger, T., Cox, J. & Mann, M. Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics 9, 2252–2261 (2010).
Liu, H., Sadygov, R.G. & Yates, J.R. III. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 76, 4193–4201 (2004).
Li, G.-Z. et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 (2009).
Michalski, A., Cox, J. & Mann, M. More than 100,000 detectable peptide species elute in single shotgun proteomics runs but the majority is inaccessible to data-dependent LC-MS/MS. J. Proteome Res. 10, 1785–1793 (2011).
Gatto, L. et al. Testing and validation of computational methods for mass spectrometry. J. Proteome Res. 15, 809–814 (2016).
Dufresne, C. et al. ABRF research group development and characterization of a proteomics normalization standard consisting of 1,000 stable isotope labeled peptides. J. Biomol. Tech. 25, S1 (2014).
Yates, J.R. III et al. Toward objective evaluation of proteomic algorithms. Nat. Methods 9, 455–456 (2012).
Leprevost, Fda.V., Barbosa, V.C., Francisco, E.L., Perez-Riverol, Y. & Carvalho, P.C. On best practices in the development of bioinformatics software. Front. Genet. 5, 199 (2014).
Pak, H. et al. Clustering and filtering tandem mass spectra acquired in data-independent mode. J. Am. Soc. Mass Spectrom. 24, 1862–1871 (2013).
The difficulty of a fair comparison. Nat. Methods 12, 273 (2015).
Kuharev, J., Navarro, P., Distler, U., Jahn, O. & Tenzer, S. In-depth evaluation of software tools for data-independent acquisition based label-free quantification. Proteomics 15, 3140–3151 (2015).
Sajic, T., Liu, Y. & Aebersold, R. Using data-independent, high-resolution mass spectrometry in protein biomarker research: perspectives and clinical applications. Proteomics Clin. Appl. 9, 307–321 (2015).
Röst, H.L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Bruderer, R. et al. Extending the limits of quantitative proteome profiling with data-independent acquisition and application to acetaminophen-treated three-dimensional liver microtissues. Mol. Cell. Proteomics 14, 1400–1410 (2015).
Reiter, L. et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat. Methods 8, 430–435 (2011).
Tsou, C.-C. et al. DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics. Nat. Methods 12, 258–264, 7, 264 (2015).
Cox, J. et al. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 13, 2513–2526 (2014).
Elias, J.E. & Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Navarro, P. et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 13, 1234–1247 (2014).
Bell, A.W. et al. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat. Methods 6, 423–430 (2009).
Schubert, O.T. et al. Building high-quality assay libraries for targeted analysis of SWATH MS data. Nat. Protoc. 10, 426–441 (2015).
Rosenberger, G. et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Sci. Data 1, 140031 (2014).
Shteynberg, D., Nesvizhskii, A.I., Moritz, R.L. & Deutsch, E.W. Combining results of multiple search engines in proteomics. Mol. Cell. Proteomics 12, 2383–2393 (2013).
Yuan, Z.-F., Lin, S., Molden, R.C. & Garcia, B.A. Evaluation of proteomic search engines for the analysis of histone modifications. J. Proteome Res. 13, 4470–4478 (2014).
Distler, U. et al. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat. Methods 11, 167–170 (2014).
Fonslow, B.R. et al. Digestion and depletion of abundant proteins improves proteomic coverage. Nat. Methods 10, 54–56 (2013).
Wis´niewski, J.R., Zougman, A., Nagaraj, N. & Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 (2009).
Escher, C. et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics 12, 1111–1121 (2012).
Eng, J.K., Jahan, T.A. & Hoopmann, M.R. Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013).
Perkins, D.N., Pappin, D.J., Creasy, D.M. & Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Reiter, L. et al. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol. Cell. Proteomics 8, 2405–2417 (2009).
Lam, H. et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics 7, 655–667 (2007).
Deutsch, E.W. et al. TraML–a standard format for exchange of selected reaction monitoring transition lists. Mol. Cell. Proteomics 11, R111.015040 (2012).
Kunszt, P. et al. iPortal: the swiss grid proteomics portal: Requirements and new features based on experience and usability considerations. Concurr. Comput. 27, 433–445 (2015).
Ning, K., Fermin, D. & Nesvizhskii, A.I. Comparative analysis of different label-free mass spectrometry based protein abundance estimates and their correlation with RNA-Seq gene expression data. J. Proteome Res. 11, 2261–2271 (2012).
Röst, H.L. et al. TRIC: an automated alignment strategy for reproducible protein quantification in targeted proteomics. Nat. Methods 13, 777–783 (2016).
Fenyö, D. & Beavis, R.C. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Anal. Chem. 75, 768–774 (2003).
Kim, S. & Pevzner, P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 5, 5277 (2014).
Nesvizhskii, A.I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Shteynberg, D. et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690 (2011).
Silva, J.C., Gorenstein, M.V., Li, G.-Z., Vissers, J.P.C. & Geromanos, S.J. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 (2006).
Vizcaíno, J.A. et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41, D1063–D1069 (2013).
Acknowledgements
We thank R. Spohrer for sample preparation and L. Burton, A. Lau and G. Ivosev for their support with SWATH 2.0. P.N. and S.T. are supported by grants from the Bundesministerium für Bildung und Forschung (BMBF) (Express2Present 0316179C), the Deutsche Forschungsgemeinschaft (DFG) (ST599/1-1, ST599/2-1) and Mainz University (Research Center for Immunotherapy (FZI)). H.L.R. is supported by Swiss National Science Foundation grant P2EZP3_162268. Y.P.-R. is supported by the Biotechnology and Biological Sciences Research Council (BBSRC) 'PROCESS' grant (BB/K01997X/1). A.I.N. is supported by US National Institutes of Health grant 5R01GM94231. R.A. was supported by European Research Council (ERC) AdG 233226 (Proteomics version 3.0) and ERC-2014-AdG 670821 (Proteomicxs 4D), the PhosphonetX project of SystemsX.ch and the Swiss National Science Foundation grant 3100A_166435.
Author information
Authors and Affiliations
Contributions
P.N. and S.T. designed and supervised the study; U.D. and L.C.G. prepared the samples and performed the MS measurements; L.C.G., G.R. and H.L.R. executed and supervised the OpenSWATH analyses; P.N. and S.A.T. executed and supervised the SWATH 2.0 analyses; P.N. and B.M. executed and supervised the Skyline analyses; P.N., O.M.B. and L.R. executed and supervised the Spectronaut analyses; C.-C.T. and A.I.N. executed and supervised the DIA-Umpire analyses; J.K., P.N. and Y.P.-R. developed LFQbench; P.N., S.T., J.K., B.M. and O.M.B. performed the benchmark analyses; L.C.G., O.M.B., B.M., H.L.R., S.A.T., C.-C.T., L.R., G.R., A.I.N. and R.A. provided critical input into the project; P.N., J.K. and S.T. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
S.A.T. is employed by SCIEX, and O.M.B. and L.R. are employed by Biognosys AG.
Supplementary information
Supplementary Text and Figures
Supplementary Tables 1–9, Supplementary Figures 1–25 and Supplementary Notes 1–6 (PDF 28116 kb)
Supplementary Code
LFQbench (R-package) source code (ZIP 19693 kb)
Supplementary Data Set 1
List of peptides exclusively identified by DIA-Umpire in the HYE124 64 variable windows experiment (XLSX 131 kb)
Supplementary Data Set 2
Ion library for 32 fixed SWATH windows experiments (ZIP 14442 kb)
Supplementary Data Set 3
Ion library for 32 variable SWATH windows experiments (ZIP 15376 kb)
Supplementary Data Set 4
Ion library for 64 fixed SWATH windows experiments (ZIP 15413 kb)
Supplementary Data Set 5
Ion library for 64 variable SWATH windows experiments (ZIP 14695 kb)
Rights and permissions
About this article
Cite this article
Navarro, P., Kuharev, J., Gillet, L. et al. A multicenter study benchmarks software tools for label-free proteome quantification. Nat Biotechnol 34, 1130–1136 (2016). https://doi.org/10.1038/nbt.3685
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3685
This article is cited by
-
SeFilter-DIA: Squeeze-and-Excitation Network for Filtering High-Confidence Peptides of Data-Independent Acquisition Proteomics
Interdisciplinary Sciences: Computational Life Sciences (2024)
-
Achieving quantitative reproducibility in label-free multisite DIA experiments through multirun alignment
Communications Biology (2023)
-
Increasing the throughput of sensitive proteomics by plexDIA
Nature Biotechnology (2023)
-
Initial recommendations for performing, benchmarking and reporting single-cell proteomics experiments
Nature Methods (2023)
-
Data-independent acquisition boosts quantitative metaproteomics for deep characterization of gut microbiota
npj Biofilms and Microbiomes (2023)