Abstract
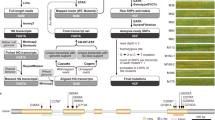
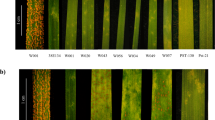
Wild relatives of domesticated crop species harbor multiple, diverse, disease resistance (R) genes that could be used to engineer sustainable disease control. However, breeding R genes into crop lines often requires long breeding timelines of 5–15 years to break linkage between R genes and deleterious alleles (linkage drag). Further, when R genes are bred one at a time into crop lines, the protection that they confer is often overcome within a few seasons by pathogen evolution1. If several cloned R genes were available, it would be possible to pyramid R genes2 in a crop, which might provide more durable resistance1. We describe a three-step method (MutRenSeq)-that combines chemical mutagenesis with exome capture and sequencing for rapid R gene cloning. We applied MutRenSeq to clone stem rust resistance genes Sr22 and Sr45 from hexaploid bread wheat. MutRenSeq can be applied to other commercially relevant crops and their relatives, including, for example, pea, bean, barley, oat, rye, rice and maize.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
McDonald, B.A. & Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379 (2002).
Dangl, J.L., Horvath, D.M. & Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 341, 746–751 (2013).
Kuang, H., Woo, S.S., Meyers, B.C., Nevo, E. & Michelmore, R.W. Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16, 2870–2894 (2004).
Smith, S.M., Pryor, A.J. & Hulbert, S.H. Allelic and haplotypic diversity at the rp1 rust resistance locus of maize. Genetics 167, 1939–1947 (2004).
Gaut, B.S., Wright, S.I., Rizzon, C., Dvorak, J. & Anderson, L.K. Recombination: an underappreciated factor in the evolution of plant genomes. Nat. Rev. Genet. 8, 77–84 (2007).
Hulbert, S.H., Webb, C.A., Smith, S.M. & Sun, Q. Resistance gene complexes: evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312 (2001).
Jupe, F. et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544 (2013).
Andolfo, G. et al. Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 14, 120 (2014).
Henry, I.M. et al. Efficient genome-wide detection and cataloging of EMS-induced mutations using exome capture and Next-Generation Sequencing. Plant Cell 26, 1382–1397 (2014).
Periyannan, S. et al. The gene Sr33, an ortholog of barley Mla genes, encodes resistance to wheat stem rust race Ug99. Science 341, 786–788 (2013).
Gerechter-Amitai, Z.K., Wahl, I., Vardi, A. & Zohary, D. Transfer of stem rust seedling resistance from wild diploid einkorn to tetraploid durum wheat by means of a triploid hybrid bridge. Euphytica 20, 281–285 (1971).
The, T.T. Chromosome location of genes conditioning stem rust resistance transferred from diploid to hexaploid wheat. Nat. New Biol. 241, 256 (1973).
Olivera Firpo, P. et al. Phenotypic and genotypic characterization of race TKTTF of Puccinia graminis f. sp. tritici that caused a wheat stem rust epidemic in southern Ethiopia in 2013/14. Phytopathology 105, 917–928 (2015).
The, T.T. et al. in 7th International Wheat Genetics Symposium (eds. Miller, T.E. & Koebner, R.M.D.) 901–909 (Bath Press, Bath, UK, 1988).
Rouse, M.N. & Jin, Y. Stem rust resistance in A-Genome diploid relatives of wheat. Plant Dis. 95, 941–944 (2011).
Witek, K. et al. Accelerated cloning of a potato late blight–resistance gene using RenSeq and SMRT sequencing. Nat. Biotechnol. doi:10.1038/nbt.3540 (2016).
Kerber, E.R. & Dyck, P.L. Inheritance in hexaploid wheat of leaf rust resistance and other characters derived from Aegilops squarrosa. Can. J. Genet. Cytol. 11, 639–647 (1969).
Marais, G.F., Potgieter, G.F., Roux, H.S. & le Roux, J. An assessment of the variation for stem rust resistance in the progeny of a cross involving the Triticum species aestivum, turgidum and tauschii. S. Afr. J. Plant Soil 11, 15–19 (1994).
Periyannan, S. et al. Identification of a robust molecular marker for the detection of the stem rust resistance gene Sr45 in common wheat. Theor. Appl. Genet. 127, 947–955 (2014).
Steuernagel, B., Jupe, F., Witek, K., Jones, J.D. & Wulff, B.B. NLR-parser: rapid annotation of plant NLR complements. Bioinformatics 31, 1665–1667 (2015).
Saintenac, C. et al. Identification of wheat gene Sr35 that confers resistance to Ug99 stem rust race group. Science 341, 783–786 (2013).
International Wheat Genome Sequencing Consortium (IWGSC). A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345, 1251788 (2014).
Mayer, K.F. et al. A physical, genetic and functional sequence assembly of the barley genome. Nature 491, 711–716 (2012).
Finn, R.D. et al. Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230 (2014).
Fu, L., Niu, B., Zhu, Z., Wu, S. & Li, W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012).
Zhang, Z., Schwartz, S., Wagner, L. & Miller, W. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7, 203–214 (2000).
Lagudah, E.S., Appels, R., Brown, A.H.D. & McNeil, D. The molecular-genetic analysis of Triticum tauschii, the D-genome donor to hexaploid wheat. Genome 34, 375–386 (1991).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Fiume, M. et al. Savant Genome Browser 2: visualization and analysis for population-scale genomics. Nucleic Acids Res. 40, W615–W621 (2012).
Paull, J.G., Pallotta, M.A., Langridge, P. & The, T.T. RFLP markers associated with Sr22 and recombination between chromosome 7A of bread wheat and the diploid species Triticum boeoticum. Theor. Appl. Genet. 89, 1039–1045 (1994).
Khan, R.R. et al. Molecular mapping of stem and leaf rust resistance in wheat. Theor. Appl. Genet. 111, 846–850 (2005).
Kota, R., Spielmeyer, W., McIntosh, R.A. & Lagudah, E.S. Fine genetic mapping fails to dissociate durable stem rust resistance gene Sr2 from pseudo-black chaff in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 112, 492–499 (2006).
Periyannan, S.K., Bansal, U.K., Bariana, H.S., Pumphrey, M. & Lagudah, E.S. A robust molecular marker for the detection of shortened introgressed segment carrying the stem rust resistance gene Sr22 in common wheat. Theor. Appl. Genet. 122, 1–7 (2011).
Akhunov, E.D. et al. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genomics 11, 702 (2010).
Luo, M.C. et al. A 4-gigabase physical map unlocks the structure and evolution of the complex genome of Aegilops tauschii, the wheat D-genome progenitor. Proc. Natl. Acad. Sci. USA 110, 7940–7945 (2013).
Rouse, M.N. & Jin, Y. Genetics of resistance to race TTKSK of Puccinia graminis f. sp. tritici in Triticum monococcum. Phytopathology 101, 1418–1423 (2011).
Krattinger, S.G. et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323, 1360–1363 (2009).
Wang, M.B., Li, Z.Y., Matthews, P.R. & Upadhyaya, N.M. Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Hortic. 461, 401–408 (1998).
Ishida, Y., Tsunashima, M., Hiei, Y. & Komari, T. Wheat (Triticum aestivum L.) transformation using immature embryos. Methods Mol. Biol. 1223, 189–198 (2015).
Acknowledgements
This research was supported by funds from the Gatsby Charitable Foundation, UK; Two Blades Foundation, USA; Biotechnology and Biological Sciences Research Council, UK; Borlaug Global Rust Initiative (BGRI) Durable Rust Resistance in Wheat (DRRW) Project (administered by Cornell University with a grant from the Bill & Melinda Gates Foundation and the UK Department for International Development); USDA-ARS National Plant Disease Recovery System; Grains Research and Development Corporation, Australia; and a fellowship to A.H. from Universiti Putra Malaysia (UPM), Malaysia. We are grateful to colleagues in The Sainsbury Laboratory and the Two Blades Foundation for helpful discussions. This research was supported in part by the NBI Computing infrastructure for Science (CiS) group and Dan MacLean's group by providing computational infrastructure.
Author information
Authors and Affiliations
Contributions
B.S., S.K.P., I.H.-P., K.W., M.N.R., G.Y., A.H., M.A., and H.B. performed experiments. B.S., S.K.P., E.S.L. and B.B.H.W. wrote the manuscript. B.S., S.K.P., K.W., J.D.G.J., E.S.L., and B.B.H.W. contributed to the design of the study.
Corresponding authors
Ethics declarations
Competing interests
B.B.H.W., B.S., E.S.L., J.D.G.J., K.W., and S.K.P. have filed two patent applications based on this work (US patent application nos. 20150240233 and 62/200,894).
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–12 (PDF 1473 kb)
Rights and permissions
About this article
Cite this article
Steuernagel, B., Periyannan, S., Hernández-Pinzón, I. et al. Rapid cloning of disease-resistance genes in plants using mutagenesis and sequence capture. Nat Biotechnol 34, 652–655 (2016). https://doi.org/10.1038/nbt.3543
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3543
This article is cited by
-
R gene-mediated resistance in the management of plant diseases
Journal of Plant Biochemistry and Biotechnology (2024)
-
Sr65: a widely effective gene for stem rust resistance in wheat
Theoretical and Applied Genetics (2024)
-
Wheat stripe rust resistance locus YR63 is a hot spot for evolution of defence genes – a pangenome discovery
BMC Plant Biology (2023)
-
Sequencing trait-associated mutations to clone wheat rust-resistance gene YrNAM
Nature Communications (2023)
-
Effects of the quantitative trait locus qPss3 on inhibition of photoperiod sensitivity and resistance to stalk rot disease in maize
Theoretical and Applied Genetics (2023)