Abstract
The study of biliary disease has been constrained by a lack of primary human cholangiocytes. Here we present an efficient, serum-free protocol for directed differentiation of human induced pluripotent stem cells into cholangiocyte-like cells (CLCs). CLCs show functional characteristics of cholangiocytes, including bile acids transfer, alkaline phosphatase activity, γ-glutamyl-transpeptidase activity and physiological responses to secretin, somatostatin and vascular endothelial growth factor. We use CLCs to model in vitro key features of Alagille syndrome, polycystic liver disease and cystic fibrosis (CF)-associated cholangiopathy. Furthermore, we use CLCs generated from healthy individuals and patients with polycystic liver disease to reproduce the effects of the drugs verapamil and octreotide, and we show that the experimental CF drug VX809 rescues the disease phenotype of CF cholangiopathy in vitro. Our differentiation protocol will facilitate the study of biological mechanisms controlling biliary development, as well as disease modeling and drug screening.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
References
Lazaridis, K.N., Strazzabosco, M. & Larusso, N.F. The cholangiopathies: disorders of biliary epithelia. Gastroenterology 127, 1565–1577 (2004).
Murray, K.F., Carithers, R.L. Jr. & AASLD. AASLD practice guidelines: evaluation of the patient for liver transplantation. Hepatology 41, 1407–1432 (2005).
Pollheimer, M.J., Trauner, M. & Fickert, P. Will we ever model PSC? – “it's hard to be a PSC model!”. Clin. Res. Hepatol. Gastroenterol. 35, 792–804 (2011).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Robinton, D.A. & Daley, G.Q. The promise of induced pluripotent stem cells in research and therapy. Nature 481, 295–305 (2012).
Sampaziotis, F., Segeritz, C.P. & Vallier, L. Potential of human induced pluripotent stem cells in studies of liver disease. Hepatology doi:10.1002/hep.27651 (2014).
Zhao, D. et al. Derivation and characterization of hepatic progenitor cells from human embryonic stem cells. PLoS ONE 4, e6468 (2009).
Tanimizu, N., Miyajima, A. & Mostov, K.E. Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture. Mol. Biol. Cell 18, 1472–1479 (2007).
Dianat, N. et al. Generation of functional cholangiocyte-like cells from human pluripotent stem cells and HepaRG cells. Hepatology 60, 700–714 (2014).
Si-Tayeb, K., Lemaigre, F.P. & Duncan, S.A. Organogenesis and development of the liver. Dev. Cell 18, 175–189 (2010).
Rashid, S.T. et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Invest. 120, 3127–3136 (2010).
Hannan, N.R., Segeritz, C.P., Touboul, T. & Vallier, L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 8, 430–437 (2013).
Xia, X., Francis, H., Glaser, S., Alpini, G. & LeSage, G. Bile acid interactions with cholangiocytes. World J. Gastroenterol. 12, 3553–3563 (2006).
Kanno, N., LeSage, G., Glaser, S., Alvaro, D. & Alpini, G. Functional heterogeneity of the intrahepatic biliary epithelium. Hepatology 31, 555–561 (2000).
Minagawa, N., Ehrlich, B.E. & Nathanson, M.H. Calcium signaling in cholangiocytes. World J. Gastroenterol. 12, 3466–3470 (2006).
Gigliozzi, A. et al. Molecular identification and functional characterization of Mdr1a in rat cholangiocytes. Gastroenterology 119, 1113–1122 (2000).
Cízková, D., Morký, J., Micuda, S., Osterreicher, J. & Martínková, J. Expression of MRP2 and MDR1 transporters and other hepatic markers in rat and human liver and in WRL 68 cell line. Physiol. Res. 54, 419–428 (2005).
Antoniou, A. et al. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology 136, 2325–2333 (2009).
Clotman, F. et al. Control of liver cell fate decision by a gradient of TGF beta signaling modulated by Onecut transcription factors. Genes Dev. 19, 1849–1854 (2005).
Turnpenny, P.D. & Ellard, S. Alagille syndrome: pathogenesis, diagnosis and management. Eur. J. Hum. Genet. 20, 251–257 (2012).
Bray, S.J. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 (2006).
Saravanamuthu, S.S., Gao, C.Y. & Zelenka, P.S. Notch signaling is required for lateral induction of Jagged1 during FGF-induced lens fiber differentiation. Dev. Biol. 332, 166–176 (2009).
Geisler, F. & Strazzabosco, M. Emerging roles of Notch signaling in liver disease. Hepatology 61, 382–392 (2015).
Zong, Y. et al. Notch signaling controls liver development by regulating biliary differentiation. Development 136, 1727–1739 (2009).
Raynaud, P. et al. A classification of ductal plate malformations based on distinct pathogenic mechanisms of biliary dysmorphogenesis. Hepatology 53, 1959–1966 (2011).
Chandok, N. Polycystic liver disease: a clinical review. Ann. Hepatol. 11, 819–826 (2012).
Temmerman, F. et al. Systematic review: the pathophysiology and management of polycystic liver disease. Aliment. Pharmacol. Ther. 34, 702–713 (2011).
Caroli, A. et al. Reducing polycystic liver volume in ADPKD: effects of somatostatin analogue octreotide. Clin. J. Am. Soc. Nephrol. 5, 783–789 (2010).
Marinelli, R.A. et al. Secretin induces the apical insertion of aquaporin-1 water channels in rat cholangiocytes. Am. J. Physiol. 276, G280–G286 (1999).
Gong, A.Y. et al. Somatostatin stimulates ductal bile absorption and inhibits ductal bile secretion in mice via SSTR2 on cholangiocytes. Am. J. Physiol. Cell Physiol. 284, C1205–C1214 (2003).
Caperna, T.J., Blomberg, L.A., Garrett, W.M. & Talbot, N.C. Culture of porcine hepatocytes or bile duct epithelial cells by inductive serum-free media. In Vitro Cell. Dev. Biol. Anim. 47, 218–233 (2011).
Masyuk, T.V., Masyuk, A.I., Torres, V.E., Harris, P.C. & Larusso, N.F. Octreotide inhibits hepatic cystogenesis in a rodent model of polycystic liver disease by reducing cholangiocyte adenosine 3′,5′-cyclic monophosphate. Gastroenterology 132, 1104–1116 (2007).
Rowe, S.M., Miller, S.B. & Sorscher, E.J. Cystic Fibrosis. N. Engl. J. Med. 352, 1992–2001 (2005).
Davies, J.C., Alton, E.W. & Bush, A. Cystic fibrosis. Br. Med. J. 335, 1255–1259 (2007).
Colombo, C. Liver disease in cystic fibrosis. Curr. Opin. Pulm. Med. 13, 529–536 (2007).
Staufer, K., Halilbasic, E., Trauner, M. & Kazemi-Shirazi, L. Cystic fibrosis related liver disease–another black box in hepatology. Int. J. Mol. Sci. 15, 13529–13549 (2014).
Haack, A., Aragão, G.G. & Novaes, M.R. Pathophysiology of cystic fibrosis and drugs used in associated digestive tract diseases. World J. Gastroenterol. 19, 8552–8561 (2013).
Hannan, N.R., Sampaziotis, F., Segeritz, C., Hanley, N. & Vallier, L. Generation of distal airway epithelium from multipotent human foregut stem cells. Stem Cells Dev. doi:10.1089/scd.2014.0512 (2015).
Ward, C.L. & Kopito, R.R. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J. Biol. Chem. 269, 25710–25718 (1994).
Shenoy, A. et al. Calcium-modulated chloride pathways contribute to chloride flux in murine cystic fibrosis-affected macrophages. Pediatr. Res. 70, 447–452 (2011).
Van Goor, F. et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 108, 18843–18848 (2011).
Clancy, J.P. et al. Results of a phase IIa study of VX-809, an investigational CFTR corrector compound, in subjects with cystic fibrosis homozygous for the F508del-CFTR mutation. Thorax 67, 12–18 (2012).
Yanai, M. et al. FGF signaling segregates biliary cell-lineage from chick hepatoblasts cooperatively with BMP4 and ECM components in vitro. Dev. Dyn. 237, 1268–1283 (2008).
Hannan, N.R. et al. Generation of multipotent foregut stem cells from human pluripotent stem cells. Stem Cell Reports 1, 293–306 (2013).
Du, P. & Kibbe, W. Lin SM lumi: a pipeline for processing Illumina microarray. Bioinformatics 24, 1547–1548 (2008).
Smyth, G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3, 3 (2004).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289–300 (1995).
Ramsay, J.O. & Silverman, B.W. Functional Data Analysis 2nd edn. (Springer, New York, 2006).
Acknowledgements
The authors would like to thank the Cambridge BRC hiPSCs core facility for the derivation of the cystic fibrosis hiPSC line, P.-A. Tsagkaraki for her help with the generation of the manuscript figures and statistical analyses, J. Skepper, L. Carter and the University of Cambridge Advanced Imaging Centre for their help with electron microscopy, C. McGee and the Wellcome Trust Sanger Institute for their help with microarray data processing and analysis, B. McLeod for IT support and S. Brown for technical support and advice. This work was funded by European Research Council starting grant Relieve IMDs (L.V., N.H.), the Cambridge Hospitals National Institute for Health Research Biomedical Research Center (L.V., N.H., F.S.), the Evelyn Trust (N.H.), the European Union FP7 grant TissuGEN (M.C.d.B.) and core support grant from the Wellcome Trust and UK Medical Research Council to the Wellcome Trust – Medical Research Council Cambridge Stem Cell Institute. F.S. has been supported by an Addenbrooke's Charitable Trust Clinical Research Training Fellowship and a joint UK Medical Research Council–Sparks Clinical Research Training Fellowship.
Author information
Authors and Affiliations
Contributions
F.S.: design and concept of study, execution of experiments and data acquisition, development of protocols and validation, generation, collection and interpretation of data, production of figures, manuscript writing, editing and final approval of manuscript. M.C.d.B., F.A.C.S.: technical support, execution of experiments. P.M.: bioinformatics and statistical analyses. A. Bertero: bioinformatics analyses. K.S.-P., E.S., E.M.: primary tissue provision. T.H.K., J.A.B., W.T.H.G., S.D., A. Baker, A.K., G.J.A.: critical revision of the manuscript for important intellectual content. N.R.F.H.: design and concept of study, study supervision, generation and interpretation of data, editing and final approval of manuscript. L.V.: design and concept of study, study supervision, interpretation of data, editing and final approval of manuscript.
Corresponding author
Ethics declarations
Competing interests
L.V is a founder and shareholder of DefiniGEN.
Integrated supplementary information
Supplementary Figure 1 Comparison between CLCs and hESC-derived cholangiocytes (hESC-Chol) generated by Dianat et al.
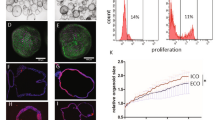
(a-d) Comparison of the transcriptional profiles of CLCs, hESC-Chol. and primary Common Bile Duct (CBD) cholangiocytes. (a, b) Hierarchical clustering (Euclidean distance, complete linkage method) of the transcriptional signatures of hESC-Chol (Dianat et al.), CBD and CLCs for (a) n= 14939 probes (b) n= 15954 genes. For each probe (a) and gene (b), standard scores (z-scores) indicating the probe (a) or gene (b) level expression relative to standard deviations above the mean for each sample were used. Color code break points were spaced equally between zmin=-0.5 and zmax=1. (c, d) Heatmap showing the hierarchically clustered Spearman correlation matrix resulting from the pair-wise comparisons of probe (c) or gene (d) expression values (z-scores). CLCs show higher correlation to CBD cholangiocytes than hESC-derived cholangiocytes. (e) Percentage of overlapping functions (POF) between primary CBD cholangiocytes and the two differentiation protocols, hepatoblast or hIPSCs for top 1000 expression-based ranked genes. Number of significant GO terms (Bonferroni adjusted p < 0.05): CLCs: 545; Primary: 449; hESC-Chol: 527; HBs: 521; hPSC: 488. (f) Percentage of CLCs responding to calcium stimulation through ATP or acetylcholine, n=4. Error bars represent SD. (g) Percentage of CLC organoids responding to secretin or somatostatin stimulation, n=4. Error bars represent SD.
Supplementary Figure 2 Generation of hepatocyte-like cells from iPSC-derived bipotent hepatoblasts.
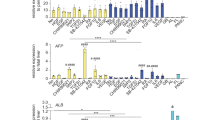
(a) IF analyses demonstrating the expression of key hepatocyte markers as indicated. Scale bars correspond to 100μm. (b) QPCR analyses demonstrating the expression of genes characteristic for hepatocytes. Center line, median; box, interquartile range; whiskers, range. Asterisks represent statistically significant differences (2-tailed t-test, ***P<0.001, ****P<0.0001). (c) Hepatocytes differentiated from iPS derived bipotent hepatoblasts exhibit cytochrome P450 activity, characteristic of primary human hepatocytes, n=9, P<0.0001 (2-tailed t-test).
Supplementary Figure 3 Activin and retinoic acid in the absence of BMP4 signaling suppress hepatic lineage commitment of bipotent hepatoblasts
(a) IF analyses demonstrating suppression of the hepatic/hepatoblast marker AFP in response to activin stimulation and the opposite effect of the activin signaling pathway inhibitor SB-431542. Scale bars correspond to 100μm. (b) IF analyses demonstrating expression of the hepatoblast (HB) marker TBX3 in response to BMP4 stimulation of bipotent hepatoblasts and the opposite effect of the BMP4 signaling pathway inhibitor noggin. Scale bars correspond to 100μm. (c) QPCR analyses demonstrating sustained expression of genes encoding hepatic and hepatoblast markers in response to BMP4 stimulation and inhibition of activin signaling through SB-431542 (SB) and suppression of the same markers in the presence of activin and retinoic acid (RA). Center line, median; box, interquartile range; whiskers, range. Asterisks represent statistically significant differences between HBs and other conditions (one-way ANOVA with Dunnett correction for multiple comparisons, *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001).
Supplementary Figure 4 Cell number expansion and differentiation efficiency for the generation of CLC organoids from hIPSCs
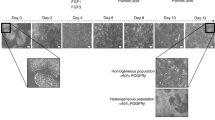
(a) Flow cytometry analyses demonstrating the percentage of cells expressing characteristic markers for key stages of CLC differentiation. The final population of CLCs is characterized in 3 lines (bottom row): A1ATD-1, FSPS11B and CF2. (b) Number of cells expressing appropriate markers for each key stage of CLC differentiation, demonstrating the generation of 57.28x106 mature (CK7+Sox9+) CLCs from a starting population of 1x106 hIPSCs. Error bars represent SD, n=3.
Supplementary Figure 5 The method for differentiation of iPSCs to CLCs is efficient across different lines.
(a) IF analyses demonstrating the expression of key biliary markers in the hIPSC line FSPS11B. Scale bar correspond to 100μm (b) QPCR analyses demonstrating the expression genes encoding early and mature biliary markers in CLCs derived from FSPS11B hIPSC-cells. Asterisks represent statistical significance (2-tailed t-test, ***P<0.001, ****P<0.0001), n=e, error bars represent SD. (c) GGT activity of CLCs derived from FSPS11B-hIPSCs cells. Center line, median; box, interquartile range; whiskers, range. Asterisks represent statistical significance (one-way ANOVA with Dunnett correction for multiple comparisons, ****P<0.0001), n=3. The data shown are complementary to figures 1,2,5 and 6 demonstrating the characterization of CLC organoids.
Supplementary Figure 6 Expression of key markers during CLC differentiation.
Heatmap demonstrating the expression of selected genes in hIPSCs, hepatoblasts (HBs), CLCs and Common Bile Duct cholangiocytes (CBD). For each probe, standard scores (z-scores) indicate the differential expression measured in number of standard deviations from the average level across all the samples.
Supplementary Figure 7 CLCs express the ileal bile acid transporter (iSBT/ASBT) but not the bile salt export pump (BSEP) physiologically expressed in hepatocytes.
(a) QPCR analyses for the expression of ASBT and BSEP in CLCs and hIPSC-derived hepatocyte-like cells (HLCs). Center line, median; box, interquartile range; whiskers, range. Asterisks represent statistical significance (one-way ANOVA with Tuckey correction for multiple comparisons, ***P<0.001, ****P<0.0001), n=3. (b) IF analyses demonstrating the expression of ASBT on the luminal surface of a cystic organoid. Scale bars: 100μM, L: Lumen
Supplementary Figure 8 The Notch signaling pathway is activated during the hepatoblast (HB) to cholangiocyte progenitor (CP) transition.
IF analyses demonstrating nuclear localization of the cleaved Notch Intracellular Domain (NICD) in CPs.
Supplementary Figure 9 Validation of PLD-hIPSCs.
(a) Sanger sequencing of PLD-hIPSCs demonstrating the presence of 2 heterozygous mutations in the PKHD1 gene.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–9, Supplementary Tables 3–6, Supplementary Note and Supplementary Methods (PDF 2880 kb)
Supplementary Table 1
Raw gene expression data (CT values) corresponding to the QPCR analyses in Figures 1d, 5f and 6a (XLSX 27 kb)
Supplementary Table 2
Microarray gene expression data corresponding to the heat map in Figure 2e (XLSX 965 kb)
Supplementary Table 7
Statistical analysis information (test statistic, degrees of freedom, statistical significance for each test) (XLSX 20 kb)
Rights and permissions
About this article
Cite this article
Sampaziotis, F., Cardoso de Brito, M., Madrigal, P. et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol 33, 845–852 (2015). https://doi.org/10.1038/nbt.3275
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.3275
This article is cited by
-
Cholangiocyte organoids to study drug-induced injury
Stem Cell Research & Therapy (2024)
-
Efficient and reproducible generation of human induced pluripotent stem cell-derived expandable liver organoids for disease modeling
Scientific Reports (2023)
-
Liver Organoids, Novel and Promising Modalities for Exploring and Repairing Liver Injury
Stem Cell Reviews and Reports (2023)
-
Genetics, pathobiology and therapeutic opportunities of polycystic liver disease
Nature Reviews Gastroenterology & Hepatology (2022)
-
Development of a 48-Well Dynamic Suspension Culture System for Pancreatic Differentiation from Human Embryonic Stem Cells
Stem Cell Reviews and Reports (2022)