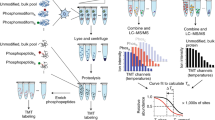
Abstract
Changes in protein conformation can affect protein function, but methods to probe these structural changes on a global scale in cells have been lacking. To enable large-scale analyses of protein conformational changes directly in their biological matrices, we present a method that couples limited proteolysis with a targeted proteomics workflow. Using our method, we assessed the structural features of more than 1,000 yeast proteins simultaneously and detected altered conformations for ∼300 proteins upon a change of nutrients. We find that some branches of carbon metabolism are transcriptionally regulated whereas others are regulated by enzyme conformational changes. We detect structural changes in aggregation-prone proteins and show the functional relevance of one of these proteins to the metabolic switch. This approach enables probing of both subtle and pronounced structural changes of proteins on a large scale.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Mischerikow, N. & Heck, A.J. Targeted large-scale analysis of protein acetylation. Proteomics 11, 571–589 (2011).
Olsen, J.V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Goh, C.S., Milburn, D. & Gerstein, M. Conformational changes associated with protein-protein interactions. Curr. Opin. Struct. Biol. 14, 104–109 (2004).
Wrabl, J.O. et al. The role of protein conformational fluctuations in allostery, function, and evolution. Biophys. Chem. 159, 129–141 (2011).
Rabiller, M. et al. Proteus in the world of proteins: conformational changes in protein kinases. Arch. Pharm. (Weinheim) 343, 193–206 (2010).
Fink, A.L. Natively unfolded proteins. Curr. Opin. Struct. Biol. 15, 35–41 (2005).
Korn, E.D., Carlier, M.F. & Pantaloni, D. Actin polymerization and ATP hydrolysis. Science 238, 638–644 (1987).
Cox, S., Radzio-Andzelm, E. & Taylor, S.S. Domain movements in protein kinases. Curr. Opin. Struct. Biol. 4, 893–901 (1994).
Bu, Z. & Callaway, D.J. Proteins move! Protein dynamics and long-range allostery in cell signaling. Adv. Protein Chem. Struct. Biol. 83, 163–221 (2011).
Chiti, F. & Dobson, C.M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 75, 333–366 (2006).
Eftink, M.R. Fluorescence techniques for studying protein structure. Methods Biochem. Anal. 35, 127–205 (1991).
Ilari, A. & Savino, C. Protein structure determination by x-ray crystallography. Methods Mol. Biol. 452, 63–87 (2008).
Pelton, J.T. & McLean, L.R. Spectroscopic methods for analysis of protein secondary structure. Anal. Biochem. 277, 167–176 (2000).
Heyduk, T. Measuring protein conformational changes by FRET/LRET. Curr. Opin. Biotechnol. 13, 292–296 (2002).
Sakakibara, D. et al. Protein structure determination in living cells by in-cell NMR spectroscopy. Nature 458, 102–105 (2009).
Herzog, F. et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science 337, 1348–1352 (2012).
Fontana, A. et al. Probing protein structure by limited proteolysis. Acta Biochim. Pol. 51, 299–321 (2004).
Aceto, A. et al. Analysis by limited proteolysis of domain organization and GSH-site arrangement of bacterial glutathione transferase B1–1. Int. J. Biochem. Cell Biol. 27, 1033–1041 (1995).
Dieckmann, R., Pavela-Vrancic, M., von Dohren, H. & Kleinkauf, H. Probing the domain structure and ligand-induced conformational changes by limited proteolysis of tyrocidine synthetase 1. J. Mol. Biol. 288, 129–140 (1999).
Peng, Z.Y. & Kim, P.S. A protein dissection study of a molten globule. Biochemistry 33, 2136–2141 (1994).
Polverino de Laureto, P. et al. Protein aggregation and amyloid fibril formation by an SH3 domain probed by limited proteolysis. J. Mol. Biol. 334, 129–141 (2003).
Picotti, P., Bodenmiller, B., Mueller, L.N., Domon, B. & Aebersold, R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795–806 (2009).
Uversky, V.N. Neuropathology, biochemistry, and biophysics of alpha-synuclein aggregation. J. Neurochem. 103, 17–37 (2007).
De Franceschi, G. et al. Molecular insights into the interaction between alpha-synuclein and docosahexaenoic acid. J. Mol. Biol. 394, 94–107 (2009).
Vilar, M. et al. The fold of alpha-synuclein fibrils. Proc. Natl. Acad. Sci. USA 105, 8637–8642 (2008).
Kendrew, J.C. et al. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature 181, 662–666 (1958).
Eliezer, D. & Wright, P.E. Is apomyoglobin a molten globule? Structural characterization by NMR. J. Mol. Biol. 263, 531–538 (1996).
Chechik, G. et al. Activity motifs reveal principles of timing in transcriptional control of the yeast metabolic network. Nat. Biotechnol. 26, 1251–1259 (2008).
DeRisi, J.L., Iyer, V.R. & Brown, P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686 (1997).
Ghaemmaghami, S. et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003).
Zhang, Q.C. et al. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature 490, 556–560 (2012).
Apweiler, R. et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 32, D115–D119 (2004).
Costenoble, R. et al. Comprehensive quantitative analysis of central carbon and amino-acid metabolism in Saccharomyces cerevisiae under multiple conditions by targeted proteomics. Mol. Syst. Biol. 7, 464 (2011).
Flikweert, M.T. et al. Pyruvate decarboxylase: an indispensable enzyme for growth of Saccharomyces cerevisiae on glucose. Yeast 12, 247–257 (1996).
Xu, Y.F. et al. Regulation of yeast pyruvate kinase by ultrasensitive allostery independent of phosphorylation. Mol. Cell 48, 52–62 (2012).
Kirtley, M.E. & McKay, M. Fructose-1,6-bisphosphate, a regulator of metabolism. Mol. Cell. Biochem. 18, 141–149 (1977).
Halfmann, R. & Lindquist, S. Epigenetics in the extreme: prions and the inheritance of environmentally acquired traits. Science 330, 629–632 (2010).
Veisova, D. et al. The C-terminal segment of yeast BMH proteins exhibits different structure compared to other 14–3-3 protein isoforms. Biochemistry 49, 3853–3861 (2010).
Thakur, S.S. et al. Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110 003699 (2011).
Kleifeld, O. et al. Isotopic labeling of terminal amines in complex samples identifies protein N-termini and protease cleavage products. Nat. Biotechnol. 28, 281–288 (2010).
Amemiya, T., Koike, R., Fuchigami, S., Ikeguchi, M. & Kidera, A. Classification and annotation of the relationship between protein structural change and ligand binding. J. Mol. Biol. 408, 568–584 (2011).
Hwang, H., Vreven, T., Janin, J. & Weng, Z. Protein-protein docking benchmark version 4.0. Proteins 78, 3111–3114 (2010).
Zacharias, M. Accounting for conformational changes during protein-protein docking. Curr. Opin. Struct. Biol. 20, 180–186 (2010).
Narayanaswamy, R. et al. Widespread reorganization of metabolic enzymes into reversible assemblies upon nutrient starvation. Proc. Natl. Acad. Sci. USA 106, 10147–10152 (2009).
Oliveira, A.P. et al. Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 8, 623 (2012).
Lomenick, B. et al. Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 106, 21984–21989 (2009).
LeVine, H. III. Quantification of beta-sheet amyloid fibril structures with thioflavin T. Methods Enzymol. 309, 274–284 (1999).
Antonini, E. & Brunori, M. Hemoglobin and Myoglobin in their Reactions with Ligands (North-Holland Pub. Co., Amsterdam, 1971).
Teale, F.W. Cleavage of the haem-protein link by acid methylethylketone. Biochim. Biophys. Acta 35, 543 (1959).
Leibundgut, M., Jenni, S., Frick, C. & Ban, N. Structural basis for substrate delivery by acyl carrier protein in the yeast fatty acid synthase. Science 316, 288–290 (2007).
Lozza, J. Crystallographic and Biochemical Studies of Fungal Fatty Acid Synthase in Complex with Flavonol Inhibitors. PhD thesis, ETH Zurich (2010).
Sherman, F. Getting started with yeast. Methods Enzymol. 350, 3–41 (2002).
Elias, J.E. & Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Heintz, D. et al. Phosphoproteome exploration reveals a reformatting of cellular processes in response to low sterol biosynthetic capacity in Arabidopsis. J. Proteome Res. 11, 1228–1239 (2012).
Xie, X. et al. A comparative phosphoproteomic analysis of a human tumor metastasis model using a label-free quantitative approach. Electrophoresis 31, 1842–1852 (2010).
Chambers, M.C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918–920 (2012).
Glatter, T. et al. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J. Proteome Res. 11, 5145–5156 (2012).
MacLean, B. et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Choi, M. et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 30, 2524–2526 (2014).
Storey, J.D. A direct approach to false discovery rates. J. R. Stat. Soc. Series B Stat. Methodol. 64, 479–498 (2002).
Fendt, S.M. et al. Tradeoff between enzyme and metabolite efficiency maintains metabolic homeostasis upon perturbations in enzyme capacity. Mol. Syst. Biol. 6, 356 (2010).
Winston, F., Dollard, C. & Ricupero-Hovasse, S.L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11, 53–55 (1995).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Berman, H.M. et al. The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000).
Pieper, U. et al. ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 39, D465–D474 (2011).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
Crooks, G.E., Hon, G., Chandonia, J.M. & Brenner, S.E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Acknowledgements
P.P. is supported by a 'Foerderungsprofessur' grant from the Swiss National Science Foundation (grant PP00P3_133670), by an EU Seventh Framework Program Reintegration grant (FP7-PEOPLE-2010-RG-277147) and by a Promedica Stiftung (grant 2-70669-11). Y.F. is supported by an ETH Research Grant (grant 4412-1); M.S. is supported by a Natural Sciences and Engineering Research Council of Canada Postgraduate Scholarship D award. G.D.F. is supported by a post-doctoral fellowship of the University of Padua. A.P.O. is supported by the SystemsX.ch project YeastX. We thank R. Costenoble, K. Kochanowski and U. Sauer (ETH Zurich) for insightful discussions and for the measurements of the intracellular concentrations of FBP and M. Peter for access to plasmid and strain collections. We are grateful to O. Vitek and M. Choi (Purdue University), R. Riek, C. Chi and P. Navarro (ETH Zurich) for helpful discussions. We also thank P. Nanni and R. Schlapbach from the Functional Genomics Centre Zurich for access to mass spectrometry instrumentation, F. Allain for access to the D-BIOL Biomolecular NMR Spectroscopy Platform at the ETH Zurich, N. Ban and M.A. Leibundgut for providing a sample of purified yeast fatty acid synthase.
Author information
Authors and Affiliations
Contributions
P.P. conceived and supervised the project. Y.F., G.D.F. and P.P. designed and performed the experiments. Y.F., G.D.F., M.S. and A.M. performed experiments and analyzed the data. P.B. contributed to mass spectrometry measurements. P.P.d.L. supervised parts of the project. A.K. analyzed the data. A.P.O. and Y.N. contributed to the analysis and validation of the metabolic data. P.P., Y.F., G.D.F. and A.K. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
P.P., Y.F. and G.D.F. are inventors on a patent application that pertains to the method presented in this study.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10, Supplementary Tables 2–6 and Supplementary Note (PDF 4522 kb)
Supplementary Table 1
Complete set of SRM assays used in this study (XLSX 514 kb)
Supplementary Table 7
LiP sites identified from spectral count cata (XLSX 664 kb)
Supplementary Table 8
Structural properties of proteins subject to LiP cleavage in the proteome of yeast grown in glucose-based medium (XLSX 628 kb)
Supplementary Table 9
Proteins that change conformational properties upon transition from glucose to ethanol growth conditions (XLSX 81 kb)
Supplementary Table 10
Background and target proteomes used in the functional enrichment analysis (XLSX 64 kb)
Supplementary Table 11
Changes in abundance and in the LiP pattern for metabolic enzymes for yeast grown in ethanol, relative to yeast grown in glucose (XLSX 21 kb)
Supplementary Table 12
Changes in the LiP pattern of TCA cycle enzymes (XLSX 43 kb)
Supplementary Table 13
Changes in the LiP pattern of pyruvate kinase (XLSX 15 kb)
Supplementary Table 14
Reversion of the proteolytic pattern of Cdc19 by addition of FBP to the extract from cells grown in ethanol (XLSX 14 kb)
Supplementary Table 15
Changes in LiP pattern of proteins in the proteome of yeast grown in ethanol upon administration of FBP (XLSX 44 kb)
Supplementary Table 16
Changes in the LiP pattern of fatty acid synthase subunits 1 and 2 (Fas1 and Fas2) upon FBP administration (XLSX 82 kb)
Supplementary Table 17
Changes in the LiP pattern of Bmh1 and Bmh2 (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Feng, Y., De Franceschi, G., Kahraman, A. et al. Global analysis of protein structural changes in complex proteomes. Nat Biotechnol 32, 1036–1044 (2014). https://doi.org/10.1038/nbt.2999
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2999
This article is cited by
-
Chemoproteomic mapping of the glycolytic targetome in cancer cells
Nature Chemical Biology (2023)
-
Elovanoid-N34 modulates TXNRD1 key in protection against oxidative stress-related diseases
Cell Death & Disease (2023)
-
How synonymous mutations alter enzyme structure and function over long timescales
Nature Chemistry (2023)
-
Universal protein misfolding intermediates can bypass the proteostasis network and remain soluble and less functional
Nature Communications (2022)
-
The emerging role of mass spectrometry-based proteomics in drug discovery
Nature Reviews Drug Discovery (2022)