Abstract
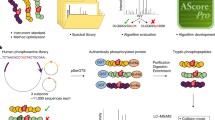
We present a peptide library and data resource of >100,000 synthetic, unmodified peptides and their phosphorylated counterparts with known sequences and phosphorylation sites. Analysis of the library by mass spectrometry yielded a data set that we used to evaluate the merits of different search engines (Mascot and Andromeda) and fragmentation methods (beam-type collision-induced dissociation (HCD) and electron transfer dissociation (ETD)) for peptide identification. We also compared the sensitivities and accuracies of phosphorylation-site localization tools (Mascot Delta Score, PTM score and phosphoRS), and we characterized the chromatographic behavior of peptides in the library. We found that HCD identified more peptides and phosphopeptides than did ETD, that phosphopeptides generally eluted later from reversed-phase columns and were easier to identify than unmodified peptides and that current computational tools for proteomics can still be substantially improved. These peptides and spectra will facilitate the development, evaluation and improvement of experimental and computational proteomic strategies, such as separation techniques and the prediction of retention times and fragmentation patterns.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chen, Y., Kwon, S.W., Kim, S.C. & Zhao, Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J. Proteome Res. 4, 998–1005 (2005).
Chen, Y., Zhang, J., Xing, G. & Zhao, Y. Mascot-derived false positive peptide identifications revealed by manual analysis of tandem mass spectra. J. Proteome Res. 8, 3141–3147 (2009).
Keller, A. et al. Experimental protein mixture for validating tandem mass spectral analysis. OMICS 6, 207–212 (2002).
Rudnick, P.A., Wang, Y., Evans, E., Lee, C.S. & Balgley, B.M. Large scale analysis of MASCOT results using a Mass Accuracy-based THreshold (MATH) effectively improves data interpretation. J. Proteome Res. 4, 1353–1360 (2005).
Klimek, J. et al. The standard protein mix database: a diverse data set to assist in the production of improved Peptide and protein identification software tools. J. Proteome Res. 7, 96–103 (2008).
Mallick, P. et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 (2007).
Nesvizhskii, A.I., Vitek, O. & Aebersold, R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat. Methods 4, 787–797 (2007).
Mallick, P. & Kuster, B. Proteomics: a pragmatic perspective. Nat. Biotechnol. 28, 695–709 (2010).
Elias, J.E. & Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 4, 207–214 (2007).
Keller, A., Nesvizhskii, A.I., Kolker, E. & Aebersold, R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 (2002).
Nesvizhskii, A.I., Keller, A., Kolker, E. & Aebersold, R. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 (2003).
Shteynberg, D. et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690 (2011).
Bohrer, B.C. et al. Combinatorial libraries of synthetic peptides as a model for shotgun proteomics. Anal. Chem. 82, 6559–6568 (2010).
Beausoleil, S.A., Villen, J., Gerber, S.A., Rush, J. & Gygi, S.P. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 (2006).
Bailey, C.M. et al. SLoMo: automated site localization of modifications from ETD/ECD mass spectra. J. Proteome Res. 8, 1965–1971 (2009).
Lemeer, S. et al. Phosphorylation site localization in peptides by MALDI MS/MS and the Mascot Delta Score. Anal. Bioanal. Chem. 402, 249–260 (2012).
Savitski, M.M. et al. Confident phosphorylation site localization using the Mascot Delta Score. Mol. Cell. Proteomics 10, M110.003830 (2011).
Taus, T. et al. Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 (2011).
Daub, H. et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 (2008).
Olsen, J.V. et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 (2006).
Olsen, J.V. et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3 (2010).
Oppermann, F.S. et al. Large-scale proteomics analysis of the human kinome. Mol. Cell. Proteomics 8, 1751–1764 (2009).
Rikova, K. et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007).
Steen, H., Jebanathirajah, J.A., Rush, J., Morrice, N. & Kirschner, M.W. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell Proteomics 5, 172–181 (2006).
Krokhin, O.V. Sequence-specific retention calculator. Algorithm for peptide retention prediction in ion-pair RP-HPLC: application to 300- and 100-A pore size C18 sorbents. Anal. Chem. 78, 7785–7795 (2006).
Jedrychowski, M.P. et al. Evaluation of HCD- and CID-type fragmentation within their respective detection platforms for murine phosphoproteomics. Mol. Cell. Proteomics 10, M111.009910 (2011).
Nagaraj, N., D'Souza, R.C., Cox, J., Olsen, J.V. & Mann, M. Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9, 6786–6794 (2010).
Swaney, D.L., McAlister, G.C. & Coon, J.J. Decision tree–driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959–964 (2008).
Swaney, D.L., Wenger, C.D., Thomson, J.A. & Coon, J.J. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. USA 106, 995–1000 (2009).
Boersema, P.J., Mohammed, S. & Heck, A.J. Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass. Spectrom. 44, 861–878 (2009).
Frese, C.K. et al. Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 10, 2377–2388 (2011).
Zhou, H. et al. Enhancing the identification of phosphopeptides from putative basophilic kinase substrates using Ti (IV) based IMAC enrichment. Mol. Cell. Proteomics 10, M110.006452 (2011).
Kapp, E.A. et al. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics 5, 3475–3490 (2005).
Frank, A.M. et al. Spectral archives: extending spectral libraries to analyze both identified and unidentified spectra. Nat. Methods 8, 587–591 (2011).
Huang, Y. et al. A data-mining scheme for identifying peptide structural motifs responsible for different MS/MS fragmentation intensity patterns. J. Proteome Res. 7, 70–79 (2008).
Lemeer, S. & Heck, A.J. The phosphoproteomics data explosion. Curr. Opin. Chem. Biol. 13, 414–420 (2009).
Baker, P.R., Trinidad, J.C. & Chalkley, R.J. Modification site localization scoring integrated into a search engine. Mol. Cell. Proteomics 10, M111.008078 (2011).
Chalkley, R.J. & Clauser, K.R. Modification site localization scoring: strategies and performance. Mol. Cell. Proteomics 11, 3–14 (2012).
Kelstrup, C.D., Hekmat, O., Francavilla, C. & Olsen, J.V. Pinpointing phosphorylation sites: quantitative filtering and a novel site-specific x-ion fragment. J. Proteome Res. 10, 2937–2948 (2011).
Krokhin, O.V. & Spicer, V. Peptide retention standards and hydrophobicity indexes in reversed-phase high-performance liquid chromatography of peptides. Anal. Chem. 81, 9522–9530 (2009).
Conrads, T.P., Anderson, G.A., Veenstra, T.D., Pasa-Tolic, L. & Smith, R.D. Utility of accurate mass tags for proteome-wide protein identification. Anal. Chem. 72, 3349–3354 (2000).
Moruz, L. et al. Chromatographic retention time prediction for posttranslationally modified peptides. Proteomics 12, 1151–1159 (2012).
Moruz, L., Tomazela, D. & Kall, L. Training, selection, and robust calibration of retention time models for targeted proteomics. J. Proteome Res. 9, 5209–5216 (2010).
Geromanos, S.J. et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695 (2009).
Hoaglund-Hyzer, C.S., Li, J. & Clemmer, D.E. Mobility labeling for parallel CID of ion mixtures. Anal. Chem. 72, 2737–2740 (2000).
Thingholm, T.E., Jensen, O.N. & Larsen, M.R. Analytical strategies for phosphoproteomics. Proteomics 9, 1451–1468 (2009).
Kyte, J. & Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132 (1982).
Zhou, H. et al. Robust phosphoproteome enrichment using monodisperse microsphere-based immobilized titanium (IV) ion affinity chromatography. Nat. Protoc. 8, 461–480 (2013).
Acknowledgements
This research was supported in part by the Deutsche Forschungsgemeinschaft International Research Training Group 'Regulation and Evolution of Cellular Systems' (RECESS, GRK 1563) and in part by the PRIME-XS project with the grant agreement number 262067 funded by the European Union 7th Framework Program. H.M. acknowledges the support of the Graduate School at the Technische Universität München, and the authors thank A. Hubauer for expert technical assistance.
Author information
Authors and Affiliations
Contributions
H.M., J.E.S. and B.K. designed the study. S.L., J.E.S., L.M. and S.M. performed experiments. H.M., S.L., L.M. and J.C. analyzed data. H.M., S.L., A.J.R.H., M.M. and B.K. wrote manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–23 (PDF 3226 kb)
Supplementary Table 1
Sequence, site of phosphorylation within the sequence, length and GRAVY score (Hydrophobicity) of the 851 representative sample peptides derived from the consensus of three out of the five publically available human phosphorylation data sets used in this study (XLSX 47 kb)
Supplementary Table 2
Peptide sequence, position of phosphorylation site in the sequence and Gravy score of the seed peptide synthesis of libraries used in this study. For each seed peptide sequence the final number of peptides in the library is given (XLSX 15 kb)
Supplementary Table 3
Search and classification result of HCD data aquired on a Orbitrap Velos. (XLSX 85450 kb)
Supplementary Table 4
Search and classification result of ETD-FT data aquired on a Orbitrap Velos. (XLSX 58136 kb)
Supplementary Table 5
Number of peptide identifications and phosphorylation site localizations at a given global or local false discovery rate (Mascot) (XLSX 526 kb)
Supplementary Table 6
Coefficients for the computation of local and global FDRs and FLRs (XLSX 551 kb)
Rights and permissions
About this article
Cite this article
Marx, H., Lemeer, S., Schliep, J. et al. A large synthetic peptide and phosphopeptide reference library for mass spectrometry–based proteomics. Nat Biotechnol 31, 557–564 (2013). https://doi.org/10.1038/nbt.2585
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2585
This article is cited by
-
Candidate biomarkers for treatment benefit from sunitinib in patients with advanced renal cell carcinoma using mass spectrometry-based (phospho)proteomics
Clinical Proteomics (2023)
-
Prediction of peptide mass spectral libraries with machine learning
Nature Biotechnology (2023)
-
DeepFLR facilitates false localization rate control in phosphoproteomics
Nature Communications (2023)
-
A multi-purpose, regenerable, proteome-scale, human phosphoserine resource for phosphoproteomics
Nature Methods (2022)
-
Affinity Selection from Synthetic Peptide Libraries Enabled by De Novo MS/MS Sequencing
International Journal of Peptide Research and Therapeutics (2022)