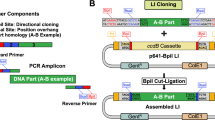
Abstract
Transcription activator–like (TAL) effector proteins derived from Xanthomonas species have emerged as versatile scaffolds for engineering DNA-binding proteins of user-defined specificity and functionality. Here we describe a rapid, simple, ligation-independent cloning (LIC) technique for synthesis of TAL effector genes. Our approach is based on a library of DNA constructs encoding individual TAL effector repeat unit combinations that can be processed to contain long, unique single-stranded DNA overhangs suitable for LIC. Assembly of TAL effector arrays requires only the combinatorial mixing of fluids and has exceptional fidelity. TAL effector nucleases (TALENs) produced by this method had high genome-editing activity at endogenous loci in HEK 293T cells (64% were active). To maximize throughput, we generated a comprehensive 5-mer TAL effector repeat unit fragment library that allows automated assembly of >600 TALEN genes in a single day. Given its simplicity, throughput and fidelity, LIC assembly will permit the generation of TAL effector gene libraries for large-scale functional genomics studies.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Boch, J. & Bonas, U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 48, 419–436 (2010).
Boch, J. et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512 (2009).
Moscou, M.J. & Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501 (2009).
Miller, J.C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Carroll, D. Genome engineering with zinc-finger nucleases. Genetics 188, 773–782 (2011).
Christian, M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010).
Li, T. et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 39, 6315–6325 (2011).
Liu, J. et al. Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J. Genet. Genomics 39, 209–215 (2012).
Tesson, L. et al. Knockout rats generated by embryo microinjection of TALENs. Nat. Biotechnol. 29, 695–696 (2011).
Huang, P. et al. Heritable gene targeting in zebrafish using customized TALENs. Nat. Biotechnol. 29, 699–700 (2011).
Sander, J.D. et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat. Biotechnol. 29, 697–698 (2011).
Wood, A.J. et al. Targeted genome editing across species using ZFNs and TALENs. Science 333, 307 (2011).
Hockemeyer, D. et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 29, 731–734 (2011).
Watanabe, T. et al. Non-transgenic genome modifications in a hemimetabolous insect using zinc-finger and TAL effector nucleases. Nat. Commun. 3, 1017 (2012).
Zhang, F. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 29, 149–153 (2011).
Weber, E., Gruetzner, R., Werner, S., Engler, C. & Marillonnet, S. Assembly of designer TAL effectors by Golden Gate cloning. PLoS ONE 6, e19722 (2011).
Morbitzer, R., Elsaesser, J., Hausner, J. & Lahaye, T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Res. 39, 5790–5799 (2011).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Geissler, R. et al. Transcriptional activators of human genes with programmable DNA-specificity. PLoS ONE 6, e19509 (2011).
Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 (2012).
Aslanidis, C. & de Jong, P.J. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 18, 6069–6074 (1990).
Geu-Flores, F., Nour-Eldin, H.H., Nielsen, M.T. & Halkier, B.A. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 35, e55 (2007).
Schmid-Burgk, J.L. et al. Rapid hierarchical assembly of medium-size DNA cassettes. Nucleic Acids Res. 40, e92 (2012).
Kim, H.J., Lee, H.J., Kim, H., Cho, S.W. & Kim, J.S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 19, 1279–1288 (2009).
Cade, L. et al. Highly efficient generation of heritable zebrafish gene mutations using homo- and heterodimeric TALENs. Nucleic Acids Res. 40, 8001–8010 (2012).
Streubel, J., Blucher, C., Landgraf, A. & Boch, J. TAL effector RVD specificities and efficiencies. Nat. Biotechnol. 30, 593–595 (2012).
Miller, J.C. et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 25, 778–785 (2007).
Acknowledgements
We thank T. Cathomen for technical advice with the T7EI assay and M. Hölzel for helpful discussion. V.H. is member of the excellence cluster ImmunoSensation and supported by grants from the German Research Foundation (SFB704 and SFB670) and the European Research Council (ERC-2009-StG 243046).
Author information
Authors and Affiliations
Contributions
J.L.S.-B., T.S. and V.H. developed the methodology, designed experiments, analyzed the data and wrote the manuscript. J.L.S.-B., T.S., V.K. and K.H. performed the experiments. V.H. supervised the project.
Corresponding author
Ethics declarations
Competing interests
J.L.S.-B., T.S. and V.H are inventors on a patent application dealing with LIC assembly of TALE genes.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–10 and Supplementary Tables 1–5, Supplementary Methods and Supplementary Note 1 (PDF 4128 kb)
Rights and permissions
About this article
Cite this article
Schmid-Burgk, J., Schmidt, T., Kaiser, V. et al. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat Biotechnol 31, 76–81 (2013). https://doi.org/10.1038/nbt.2460
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt.2460
This article is cited by
-
High-throughput process development from gene cloning to protein production
Microbial Cell Factories (2023)
-
Enhancing the quality of staple food crops through CRISPR/Cas-mediated site-directed mutagenesis
Planta (2023)
-
Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects
Signal Transduction and Targeted Therapy (2020)
-
Primordial germ cell-mediated transgenesis and genome editing in birds
Journal of Animal Science and Biotechnology (2018)
-
CRISPR/Cas9 genome editing technology significantly accelerated herpes simplex virus research
Cancer Gene Therapy (2018)