Abstract
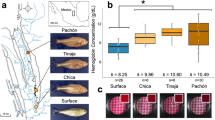
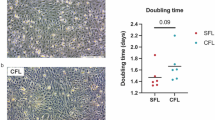
Periodic food shortages are a major challenge faced by organisms in natural habitats. Cave-dwelling animals must withstand long periods of nutrient deprivation, as—in the absence of photosynthesis—caves depend on external energy sources such as seasonal floods1. Here we show that cave-adapted populations of the Mexican tetra, Astyanax mexicanus, have dysregulated blood glucose homeostasis and are insulin-resistant compared to river-adapted populations. We found that multiple cave populations carry a mutation in the insulin receptor that leads to decreased insulin binding in vitro and contributes to hyperglycaemia. Hybrid fish from surface–cave crosses carrying this mutation weigh more than non-carriers, and zebrafish genetically engineered to carry the mutation have increased body weight and insulin resistance. Higher body weight may be advantageous in caves as a strategy to cope with an infrequent food supply. In humans, the identical mutation in the insulin receptor leads to a severe form of insulin resistance and reduced lifespan. However, cavefish have a similar lifespan to surface fish and do not accumulate the advanced glycation end-products in the blood that are typically associated with the progression of diabetes-associated pathologies. Our findings suggest that diminished insulin signalling is beneficial in a nutrient-limited environment and that cavefish may have acquired compensatory mechanisms that enable them to circumvent the typical negative effects associated with failure to regulate blood glucose levels.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
13 November 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Culver, D. C. & Pipan, T. The Biology of Caves and Other Subterranean Habitats (Oxford Univ. Press, 2009)
Horst Wilkens, U. S. Evolution in the Dark, Darwin’s Loss Without Selection (Springer, 2017)
Aspiras, A. C., Rohner, N., Martineau, B., Borowsky, R. L. & Tabin, C. J. Melanocortin 4 receptor mutations contribute to the adaptation of cavefish to nutrient-poor conditions. Proc. Natl Acad. Sci. USA 112, 9668–9673 (2015)
Moran, D., Softley, R. & Warrant, E. J. Eyeless Mexican cavefish save energy by eliminating the circadian rhythm in metabolism. PLoS ONE 9, e107877 (2014)
Hüppop, K. Oxygen consumption of Astyanax fasciatus (Characidae, Pisces): a comparison of epigean and hypogean populations. Environ. Biol. Fishes 17, 299–308 (1986)
Gross, J. B. The complex origin of Astyanax cavefish. BMC Evol. Biol. 12, 105 (2012)
Bradic, M., Teotónio, H. & Borowsky, R. L. The population genomics of repeated evolution in the blind cavefish Astyanax mexicanus. Mol. Biol. Evol. 30, 2383–2400 (2013)
Saltiel, A. R. & Kahn, C. R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 (2001)
Rines, A. K., Sharabi, K., Tavares, C. D. & Puigserver, P. Targeting hepatic glucose metabolism in the treatment of type 2 diabetes. Nat. Rev. Drug Discov. 15, 786–804 (2016)
Navarro, I. et al. Insights into insulin and glucagon responses in fish. Fish Physiol. Biochem. 27, 205–216 (2002)
Lizcano, J. M. & Alessi, D. R. The insulin signalling pathway. Curr. Biol. 12, R236–R238 (2002)
McGaugh, S. E. et al. The cavefish genome reveals candidate genes for eye loss. Nat. Commun. 5, 5307 (2014)
Atray, A. et al. Rabson Mendenhall Syndrome; a case report. J. Diabetol. 2, 2 (2013)
Carrera, P. et al. Substitution of Leu for Pro-193 in the insulin receptor in a patient with a genetic form of severe insulin resistance. Hum. Mol. Genet. 2, 1437–1441 (1993)
Taylor, S. I. et al. Mutations in insulin-receptor gene in insulin-resistant patients. Diabetes Care 13, 257–279 (1990)
Bradic, M., Beerli, P., García-de León, F. J., Esquivel-Bobadilla, S. & Borowsky, R. L. Gene flow and population structure in the Mexican blind cavefish complex (Astyanax mexicanus). BMC Evol. Biol. 12, 9 (2012)
Albadri, S., Del Bene, F. & Revenu, C. Genome editing using CRISPR/Cas9-based knock-in approaches in zebrafish. Methods 121-122, 77–85 (2017)
Savage, D. B. & Semple, R. K. Recent insights into fatty liver, metabolic dyslipidaemia and their links to insulin resistance. Curr. Opin. Lipidol. 21, 329–336 (2010)
Suzuki, N., Kitamura, K. I. & Hattori, A. Fish scale is a suitable model for analyzing determinants of skeletal fragility in type 2 diabetes. Endocrine 54, 575–577 (2016)
Simon, V. et al. Comparing growth in surface and cave morphs of the species Astyanax mexicanus: insights from scales. Evodevo 8, 23 (2017)
Hayes, A. J. et al. Spinal deformity in aged zebrafish is accompanied by degenerative changes to their vertebrae that resemble osteoarthritis. PLoS ONE 8, e75787 (2013)
Yan, S. F., Ramasamy, R. & Schmidt, A. M. Mechanisms of disease: advanced glycation end-products and their receptor in inflammation and diabetes complications. Nat. Clin. Pract. Endocrinol. Metab. 4, 285–293 (2008)
Prasad, A., Bekker, P. & Tsimikas, S. Advanced glycation end products and diabetic cardiovascular disease. Cardiol. Rev. 20, 177–183 (2012)
De Meyts, P. & Whittaker, J. Structural biology of insulin and IGF1 receptors: implications for drug design. Nat. Rev. Drug Discov. 1, 769–783 (2002)
Borowsky, R. Restoring sight in blind cavefish. Curr. Biol. 18, R23–R24 (2008)
Elipot, Y., Legendre, L., Père, S., Sohm, F. & Rétaux, S. Astyanax transgenesis and husbandry: how cavefish enters the laboratory. Zebrafish 11, 291–299 (2014)
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014)
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, https://doi.org/10.14806/ej.17.1.200 (2011)
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010)
Van der Auwera, G. A. et al. From FastQ data to high-confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinform. 43, 11.10.1–11.10.33 (2013)
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011)
Fariello, M. I., Boitard, S., Naya, H., SanCristobal, M. & Servin, B. Detecting signatures of selection through haplotype differentiation among hierarchically structured populations. Genetics 193, 929–941 (2013)
Tomomori-Sato, C. et al. A mammalian mediator subunit that shares properties with Saccharomyces cerevisiae mediator subunit Cse2. J. Biol. Chem. 279, 5846–5851 (2004)
Murphy, R. F., Powers, S., Verderame, M., Cantor, C. R. & Pollack, R. Flow cytofluorometric analysis of insulin binding and internalization by Swiss 3T3 cells. Cytometry 2, 402–406 (1982)
R Development Core Team. A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016)
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2009)
Acknowledgements
We thank Y. Chinchore and C. Sengel for technical advice; X. Gao for bioinformatics support; Z. Zakibe for photographs of the fish; the Aquatics facility at Stowers for fish maintenance and support; the cell culture core at Stowers for cell line maintenance and advice; the molecular biology core at Stowers for design, execution and validation of the CRISPR constructs; the proteomics core; M. Levy for advice and computational modelling of the insulin receptor; A. Herman for help with the genome scan; the Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School; M. Miller for illustration; and S. Williams, F. Damen, S. Xiong, E. Kingsley and K. Fox for feedback on the manuscript text. This work was supported by a grant from the NIH to C.J.T. (HD089934) and institutional funding to N.R. M.R.R. was supported by a National Research Service Award (DK108495) and R.P. was supported by a grant from the Deutsche Forschungsgemeinschaft (PE 2807/1-1).
Author information
Authors and Affiliations
Contributions
M.R.R., A.C.A., C.J.T. and N.R. conceived the project and designed research with additional contributions from K.G., R.P. and A.C.B. M.R.R., A.C.A., K.G., R.P., J.Y.S., B.M., M.P., A.C.B., J.A.T., S.M., R.B. and N.R. performed the research. M.R.R., A.C.A., C.J.T. and N.R. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Reviewer Information Nature thanks K. Kavanagh, S. O’Rahilly and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Numbers of insulin- and glucagon-positive cells in the developing pancreas are unchanged in Tinaja cavefish relative to surface fish.
a, Whole-mount immunohistochemical detection of insulin- and glucagon-positive cells in Tinaja larvae at 10 days post fertilization. b, Number of glucagon- and insulin-positive cells in surface and Tinaja larvae at 10–11 days post fertilization (n = 5 fish per population,). c, Average number of glucagon- and insulin-positive cells, fish length, ratio of insulin to glucagon positive cells and P value comparing the surface and Tinaja values (determined using Student’s t-test).
Extended Data Figure 2 Serum glucagon levels are comparable between the different populations.
Box plot comparing serum glucagon levels between surface, Tinaja, Molino and Pachón fish after 24-h fast. n = 12 fish per population, average of 57.87, 59.76, 79.66 and 48.89 respectively. P = 0.52, one-way ANOVA. Box plots show 25th, 50th and 75th percentiles (horizontal bars), and 1.5× interquartile ranges (error bars), dots represent outliers.
Extended Data Figure 3 Serum insulin levels are comparable between surface and Tinaja fish.
a, Serum blotted onto nitrocellulose membrane using Bio-Dot SF microfiltration apparatus (Bio-Rad, catalogue number 1706542) probed with anti-insulin antibody (DAKO). Each blot represents an individual fish between 1- and 2-years-old (n = 24 fish per population). b, Quantification of insulin level measured by densitometry of blots. AU, artificial units; median, 25th, 50th and 75th percentiles (horizontal bars) and error bars at 1.5× interquartile ranges. Tinaja cavefish insulin levels (mean = 10,770) tended to be higher than those of surface fish (mean = 7,194) but the results are not significant (P = 0.057, Student’s two-sample t-test).
Extended Data Figure 4 Insulin decreases blood glucose level in surface fish.
a, We injected different concentrations of human recombinant insulin (Sigma, product I9278, stock 9.5–11.5 mg ml−1) into the intraperitoneal cavity of surface fish to determine the effective dosage for subsequent experiments. Blood glucose levels are significantly lower after injection of approximately 0.6 or 0.06 g insulin per mg of fish weight compared to 0.0006 g (30 min after injection, n = 4 fish per dosage, dots represent individual fish, significance calculated using one-way ANOVA with Tukey’s HSD post hoc test, *P < 0.05). We used 0.06 g insulin per mg of fish in subsequent experiments. b, Blood glucose levels of surface fish over time, after injection of PBS or insulin. Blood glucose levels are significantly lower at 60 and 90 min compared to 15 min after insulin injection (n = 10 fish per time point and condition, significance calculated using one-way ANOVA with Tukey’s HSD post hoc test, *P < 0.05). Therefore, we focused on the 60-min time point for comparisons with Tinaja cavefish. c, Blood glucose levels at 15 and 60 min after insulin injection in surface fish and Tinaja cavefish. Surface fish display a significant decrease in blood glucose levels, whereas cavefish display a significant increase in blood glucose levels (significance calculated using two-tailed Student’s t-test, *P < 0.05, ***P < 0.0005). Tinaja cavefish blood glucose levels may increase owing to the stress of being injected; stress hormones, such as catecholamines, ACTH and epinephrine, cause transient increases in blood glucose in humans and mice, an effect that cannot be mitigated in the absence of insulin signalling. Although both cavefish and surface fish probably undergo a stress response upon injection, this is overcome in the surface fish, which have wild-type insulin activity, but not in the Tinaja cavefish, which have reduced insulin signalling.
Extended Data Figure 5 An elevated fasting blood glucose level correlates with the presence of the P211L allele in F2 hybrids.
Blood glucose levels of 192 F2 surface–Tinaja hybrids of the indicated genotype 24 h after feeding. All of the fish with elevated blood glucose (greater than 60 mg dl−1) carry the P211L allele. Median, 25th, 50th and 75th percentiles (horizontal bars), and error bars at 1.5× interquartile ranges.
Extended Data Figure 6 Egg mass is a confounding variable in female fish.
a, Image of an F2 surface–Tinaja hybrid female and removed gonad, of indicated weights. b, Histogram displaying per cent gonad weight (gonad weight/total weight, multiplied by 100) of 62 F2 surface–Tinaja hybrid females fed 6 mg per day for 4 months (minimum = 3.57, 1st quartile = 10.64, median = 13.96, mean = 13.96, 3rd quartile = 17.57 and maximum = 41.86).
Extended Data Figure 7 Genome editing strategy.
CRISPR–Cas9 mediated genome editing strategy in exon 3 of the insulin receptor a (insra) zebrafish gene. The guide RNA target sequence is emphasized in bold in both the reverse strand of the wild-type genomic DNA and in the ssODN. The intended SNP exchanges are underlined, and the specific C632T to alter the P211 to L is denoted with a star. Both ends of the ssODN are protected by three phosphorothioate bonds, denoted with asterisks. The protospacer adjacent motif is shown in orange.
Extended Data Figure 8 Scale growth is impaired in the insra zebrafish mutant.
a, Quantification of scale size in zebrafish of the indicated genotype; each point represents the mean scale size of an individual fish based on the measurement of 10–14 scales removed from the left side of the body from the posterior edge of the dorsal fin to the posterior edge of the ventral fin by gentle scraping with a scalpel. Wild type (P), n = 4, P211L mutant (L), n = 3. b, c, Representative images of scales stained with 0.005% calcein with contrast and brightness adjusted to show scale edges. Significance calculated using two-way t-test of mean values, *P < 0.05.
Supplementary information
Supplementary Figure
This file contains raw uncropped film for the western blot shown in Figure 1F. Both films were from the same set of membranes which were stripped and reprobed. Red outline denotes gels used for the figure and yellow outline shows cropping used. (PDF 4014 kb)
Supplementary Data
This file contains Sequence information of the genes from the insulin signaling pathway (http://www.genome.jp/kegg-bin/show_pathway?hsa04910) from Astyanax mexicanus in FASTA format. (HTML 14399 kb)
Supplementary Information
This file contains names of the rivers in which surface Astyanax mexicanus were sampled and the numbers of individual samples collected. (PDF 35 kb)
Supplementary Information
This file contains extended text describing the population genomic approach to test the insra locus for signatures of selection. (PDF 191 kb)
Rights and permissions
About this article
Cite this article
Riddle, M., Aspiras, A., Gaudenz, K. et al. Insulin resistance in cavefish as an adaptation to a nutrient-limited environment. Nature 555, 647–651 (2018). https://doi.org/10.1038/nature26136
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature26136
This article is cited by
-
The nature and distribution of putative non-functional alleles suggest only two independent events at the origins of Astyanax mexicanus cavefish populations
BMC Ecology and Evolution (2024)
-
Phylogenetic conservation of the interdependent homeostatic relationship of sleep regulation and redox metabolism
Journal of Comparative Physiology B (2024)
-
The gut microbiome mediates adaptation to scarce food in Coleoptera
Environmental Microbiome (2023)
-
Microbial community structure and diversity in fish-flower (mint) symbiosis
AMB Express (2023)
-
Age, growth, and energy storage of the subterranean fish Triplophysa rosa (Cypriniformes: Nemacheilidae) from Chongqing, China
BMC Ecology and Evolution (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.