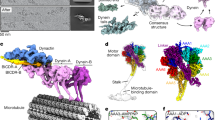
Abstract
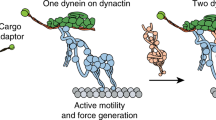
Dynein and its cofactor dynactin form a highly processive microtubule motor in the presence of an activating adaptor, such as BICD2. Different adaptors link dynein and dynactin to distinct cargoes. Here we use electron microscopy and single-molecule studies to show that adaptors can recruit a second dynein to dynactin. Whereas BICD2 is biased towards recruiting a single dynein, the adaptors BICDR1 and HOOK3 predominantly recruit two dyneins. We find that the shift towards a double dynein complex increases both the force and speed of the microtubule motor. Our 3.5 Å resolution cryo-electron microscopy reconstruction of a dynein tail–dynactin–BICDR1 complex reveals how dynactin can act as a scaffold to coordinate two dyneins side-by-side. Our work provides a structural basis for understanding how diverse adaptors recruit different numbers of dyneins and regulate the motile properties of the dynein–dynactin transport machine.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Roberts, A. J., Kon, T., Knight, P. J., Sutoh, K. & Burgess, S. A. Functions and mechanics of dynein motor proteins. Nat. Rev. Mol. Cell Biol. 14, 713–726 (2013)
Klinman, E. & Holzbaur, E. L. F. Comparative analysis of axonal transport markers in primary mammalian neurons. Methods Cell Biol. 131, 409–424 (2016)
Burkhardt, J. K., Echeverri, C. J., Nilsson, T. & Vallee, R. B. Overexpression of the dynamitin (p50) subunit of the dynactin complex disrupts dynein-dependent maintenance of membrane organelle distribution. J. Cell Biol. 139, 469–484 (1997)
Ben-Yaakov, K. et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 31, 1350–1363 (2012)
McKenney, R. J., Huynh, W., Tanenbaum, M. E., Bhabha, G. & Vale, R. D. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 345, 337–341 (2014)
Schlager, M. A., Hoang, H. T., Urnavicius, L., Bullock, S. L. & Carter, A. P. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 33, 1855–1868 (2014)
Chowdhury, S., Ketcham, S. A., Schroer, T. A. & Lander, G. C. Structural organization of the dynein-dynactin complex bound to microtubules. Nat. Struct. Mol. Biol. 22, 345–347 (2015)
Urnavicius, L. et al. The structure of the dynactin complex and its interaction with dynein. Science 347, 1441–1446 (2015)
Zhang, K. et al. Cryo-EM reveals how human cytoplasmic dynein is auto-inhibited and activated. Cell 169, 1303–1314.e18 (2017)
Redwine, W. B. et al. The human cytoplasmic dynein interactome reveals novel activators of motility. eLife 6, e28257 (2017)
Olenick, M. A., Tokito, M., Boczkowska, M., Dominguez, R. & Holzbaur, E. L. Hook adaptors induce unidirectional processive motility by enhancing the dynein–dynactin interaction. J. Biol. Chem. 291, 18239–18251 (2016)
Gama, J. B. et al. Molecular mechanism of dynein recruitment to kinetochores by the Rod–Zw10–Zwilch complex and Spindly. J. Cell Biol. 216, 943–960 (2017)
Zheng, W. Probing the energetics of dynactin filament assembly and the binding of cargo adaptor proteins using molecular dynamics simulation and electrostatics-based structural modeling. Biochemistry 56, 313–323 (2017)
Schlager, M. A. et al. Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell Reports 8, 1248–1256 (2014)
Cianfrocco, M. A., DeSantis, M. E., Leschziner, A. E. & Reck-Peterson, S. L. Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol. 31, 83–108 (2015)
Schlager, M. A. et al. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J. 29, 1637–1651 (2010)
Bielska, E. et al. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J. Cell Biol. 204, 989–1007 (2014)
Zhang, J., Qiu, R., Arst, H. N., Peñalva, M. A. & Xiang, X. HookA is a novel dynein–early endosome linker critical for cargo movement in vivo. J. Cell Biol. 204, 1009–1026 (2014)
Schroeder, C. M. & Vale, R. D. Assembly and activation of dynein–dynactin by the cargo adaptor protein Hook3. J. Cell Biol. 214, 309–318 (2016)
Grotjahn, D. A . et al. Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-018-0027-7 (2018)
Belyy, V. et al. The mammalian dynein–dynactin complex is a strong opponent to kinesin in a tug-of-war competition. Nat. Cell Biol. 18, 1018–1024 (2016)
Svoboda, K., Schmidt, C. F., Schnapp, B. J. & Block, S. M. Direct observation of kinesin stepping by optical trapping interferometry. Nature 365, 721–727 (1993)
Rai, A. K., Rai, A., Ramaiya, A. J., Jha, R. & Mallik, R. Molecular adaptations allow dynein to generate large collective forces inside cells. Cell 152, 172–182 (2013)
Rai, A. et al. Dynein clusters into lipid microdomains on phagosomes to drive rapid transport toward lysosomes. Cell 164, 722–734 (2016)
Hendricks, A. G., Holzbaur, E. L. F. & Goldman, Y. E. Force measurements on cargoes in living cells reveal collective dynamics of microtubule motors. Proc. Natl Acad. Sci. USA 109, 18447–18452 (2012)
Driller-Colangelo, A. R., Chau, K. W. L., Morgan, J. M. & Derr, N. D. Cargo rigidity affects the sensitivity of dynein ensembles to individual motor pausing. Cytoskeleton (Hoboken) 73, 693–702 (2016)
Derr, N. D. et al. Tug-of-war in motor protein ensembles revealed with a programmable DNA origami scaffold. Science 338, 662–665 (2012)
Reck-Peterson, S. L. et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell 126, 335–348 (2006)
Baumbach, J. et al. Lissencephaly-1 is a context-dependent regulator of the human dynein complex. eLife 6, e21768 (2017)
Gutierrez, P. A., Ackermann, B. E., Vershinin, M. & McKenney, R. J. Differential effects of the dynein-regulatory factor Lissencephaly-1 on processive dynein–dynactin motility. J. Biol. Chem. 292, 12245–12255 (2017)
Schroeder, C. M., Ostrem, J. M. L., Hertz, N. T. & Vale, R. D. A Ras-like domain in the light intermediate chain bridges the dynein motor to a cargo-binding region. eLife 3, e03351 (2014)
Hall, J., Song, Y., Karplus, P. A. & Barbar, E. The crystal structure of dynein intermediate chain–light chain roadblock complex gives new insights into dynein assembly. J. Biol. Chem. 285, 22566–22575 (2010)
Williams, J. C. et al. Structural and thermodynamic characterization of a cytoplasmic dynein light chain–intermediate chain complex. Proc. Natl Acad. Sci. USA 104, 10028–10033 (2007)
King, S. J., Bonilla, M., Rodgers, M. E. & Schroer, T. A. Subunit organization in cytoplasmic dynein subcomplexes. Protein Sci. 11, 1239–1250 (2002)
Tomishige, M. & Vale, R. D. Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J. Cell Biol. 151, 1081–1092 (2000)
Dodd, R. B. et al. Solution structure of the Kaposi’s sarcoma-associated herpesvirus K3 N-terminal domain reveals a novel E2-binding C4HC3-type RING domain. J. Biol. Chem. 279, 53840–53847 (2004)
Studier, F. W. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 (2005)
Incardona, M. F . et al. EDNA: a framework for plugin-based applications applied to X-ray experiment online data analysis. J. Synchrotron Radiat. 16, 872–879 (2009)
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D 67, 355–367 (2011)
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017)
Mindell, J. A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003)
Kimanius, D., Forsberg, B. O., Scheres, S. H. & Lindahl, E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife 5, e18722 (2016)
Bharat, T. A. M., Russo, C. J., Löwe, J., Passmore, L. A. & Scheres, S. H. W. Advances in single-particle electron cryomicroscopy structure determination applied to sub-tomogram averaging. Structure 23, 1743–1753 (2015)
Nguyen, T. H. D. et al. Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature 530, 298–302 (2016)
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004)
Kelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N. & Sternberg, M. J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protocols 10, 845–858 (2015)
Zhang, K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 193, 1–12 (2016)
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007)
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012)
Kucukelbir, A., Sigworth, F. J. & Tagare, H. D. Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014)
Drozdetskiy, A., Cole, C., Procter, J. & Barton, G. J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 43, W389–W394 (2015)
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014)
Acknowledgements
We thank S. Scheres, X. Bai, K. Vinothkumar and R. Leiro for cryo-EM advice; S. Chen, G. McMullan, C. Savva, G. Cannone, J. Grimmett and T. Darling for technical support; S. Bullock for SNAPf–dynein (1-1074-GST); M. Yu for crystallography support and the European Synchrotron Radiation Facility (beamline ID29) for data collection; T. Croll for model building; S. Bullock, L. Passmore, S. Lacey and H. Foster for manuscript comments; and G. Lander for discussions. This work was funded by Wellcome Trust (WT100387) and MRC grants (MC_UP_A025_1011) to A.P.C.; and NIH (GM094522) and NSF (MCB-1055017, MCB-1617028) grants to A.Y.
Author information
Authors and Affiliations
Contributions
L.U. performed all cryo-EM work on TDR, and C.K.L. performed all cryo-EM work on TDH. L.U., C.K.L., M.M.E and A.P.C. performed single-molecule experiments. L.U. performed negative-stain electron microscopy. M.M.E. and A.Y. performed optical trapping. E.M.-R. determined the NDD structure. C.M. made dynein (1-1455). A.P.C., L.U. and C.K.L. built and refined the TDR model and prepared the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Figure 1 Single particle cryo-EM analysis of TDR and TDH.
a, Cryo-EM reconstruction of the TDR complex analysed by ResMap52 and showing resolution distribution from 4 to 12 Å. b, The gold-standard Fourier shell correlation (FSC) curve of the 6.5 Å TDR map. c, Cryo-EM reconstruction of the TDH complex, showing resolution distribution from 4 to 12 Å. d, The gold-standard FSC curve of the 6.7 Å TDH map. e, Cryo-EM density for TDR low-pass filtered to 6.7 Å resolution (coloured according to cartoon) and to 20 Å (transparent outline). Density at the N terminus of BICDR1 is boxed. f, Cryo-EM density for TDH low-pass filtered to 6.7 Å (coloured according to cartoon) and to 20 Å (transparent outline) showing the putative Hook domain, an extension of the HOOK3 coiled coil ending in extra density near dynein-B (dashed box).
Extended Data Figure 2 Single-molecule assay speed distributions.
a, A one-cumulative frequency distribution plot showing run-lengths of DDB, DDR and DDH, with fit to a one-phase exponential decay. The decay constant (run length) and R2 value (least squares regression) of the fit are shown. We measured 785, 677 and 684 events for DDB, DDH and DDR, respectively, from microtubules of at least 20 μm in length from three chambers. b–f, Distribution of mean velocities of processive (unidirectional, minus-end-directed) events for DDB (n = 3,343) and DDR (n = 3,162) (b); DDB and DDH (n = 3,744) (c); active mutant dynein in complex with dynactin and BICD2 (mtDDB, n = 905) or BICDR1 (mtDDR, n = 1,183) (d); the colocalized mtDDR complexes containing both TMR–dynein tail and Alexa Fluor 647-full-length dynein (tail–dynein, n = 939) or Alexa Fluor 647-only complexes containing only full-length dynein (dynein-only, n = 1,004) (e); and all DDB complexes and complexes with both fluorophores, and hence two dyneins (colocalizers, n = 660) (f). Mean ± s.e.m. values were estimated by fitting the histograms to a Gaussian distribution (dashed lines).
Extended Data Figure 3 Single-particle cryo-EM analysis of TDR complex at 3.5 Å resolution.
a, Micrograph of the TDR complex (representative of 26,906 micrographs). b, Typical 2D-class averages of TDR in different orientations. c, The overall density map of TDR was analysed by ResMap, showing a resolution distribution from 3 to 8 Å. d, The gold-standard FSC curve of the overall TDR map. e, Mesh representation of 3.4 Å resolution density map of α helixes from dynein-B1 obtained by focused 3D classification and refinement. f, Sample density obtained by local sub-volume averaging, showing β strands from IC WD40.
Extended Data Figure 4 Cryo-EM data procedures of TDR.
Focused 3D classification and refinement procedures used in this study to improve density maps for dynein tails.
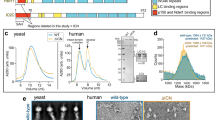
Extended Data Figure 5 Secondary structure diagram of dynein HC.
a, Secondary structure elements of dynein HC are matched against the primary sequence showing the NDD (purple) and the dynein helical bundles (blue; cyan; green; yellow; pale yellow; orange; red; pink). b, Secondary structure elements of IC. Extended N-terminal regions are coloured purple and other elements are coloured according to the blade of the WD40 domain to which they belong, except sheet β5, which associates with β30–32. c, Secondary structure elements of LIC, showing the globular domain helices and sheets (blue) and the two helices that pack against the HC (red). Jpred53 secondary structure predictions of features not seen in the electron microscopy map are shown in grey.
Extended Data Figure 6 Interactions between dynein subunits.
a, The dynein HC (yellow) interacts with the IC WD40 domain (blue) using bundles 4 and 5, with a helical segment (red cartoon) sitting in the WD40 central cavity. Dynein-A2 is shown. Interacting residues are shown as sticks (bottom panel), with HC residues in red and IC residues in green. b, Density map and model showing how the LIC (density and cartoon, blue) N- and C-terminal regions extend from the globular domain and pack against the HC (density, coloured by bundle number). Dynein-A2 is shown. c, ROBL1 (cartoon, light and dark pink) makes contacts with the IC N-terminal helices (cartoon, light and dark blue), which mediate the interaction between ROBL1 and the IC WD40 (surface). d, Representative density from the 1.9 Å resolution NDD crystal structure. e, Cartoon model of the NDD showing one chain in rainbow spectrum.
Extended Data Figure 7 Dynein–dynein contacts and interactions at the BICDR1 N terminus.
a, Conservation diagram showing sequence similarity between A2 and B1 interacting residues. Residues coloured white with red background are completely conserved, whereas residues coloured red show sequence similarity at that position. Residues at each interaction site are numbered below the alignment (A2 residues in yellow circles, B1 residues in red circles). These numbers label the accompanying cartoon to show the dynein chains that constitute each interaction. Alignment generated by ESPript54 (http://espript.ibcp.fr). b, Intermediate chain interactions showing connections between the IC of A1 and the HC of A2; the IC of A2 and the HC of B1; and the IC of B1 and the HC of B2. Interacting sites on each IC are shown as yellow spheres; sites on each HC are shown as red spheres. c, B1 (pink) contacts extra density (labelled, blue) adjacent to the BICDR1 coiled coil. The cartoon below shows the location of the area depicted (correspondingly coloured). d, Weak density connects the extra density with the LIC A2 helix 13 (blue). A cartoon representation of the area depicted is shown below.
Extended Data Figure 8 Comparison between different adaptors recruiting dynein.
a, The TDR structure (left) is compared to models of TDH (middle) and TDB (right). Although the paths of BICDR1 (yellow), HOOK3 (magenta) and BICD2 (orange) vary along the surface of dynactin (green surface), dynein-A HCs (light blue) bind at the same sites in each complex. b, Zoomed-in views of the barbed end of dynactin show that BICD2 adopts an upwards position to contact ARP1A (grey), whereas BICDR1 and HOOK3 adopt lower positions to bind dynein-B using the region coloured in red. The BICD2–ARP1A interaction site is highlighted in purple.
Supplementary information
The dynein tail/dynactin/BICDR1 complex
The full dynein tail/dynactin/BICDR1 complex (PDB:6F1T) is shown with the dynein tails (blue and red) and BICDR1 (yellow) shown in cartoon representation, and dynactin shown in surface representation (green). The complex is rotated, first to show the heavy chains in the grooves along dynactin’s filament, then to show interactions between the dynein tails and BICDR1. (MP4 12148 kb)
The cascade of dynein/dynein interactions
This video starts by focusing in on the two dynein tails from PDB 6F1T. Dynein interactions are then shown in sphere representation (colored according to Fig. 5b and Extended Data Fig. 7b). The complex is rocked to show the extensive interactions between dynein chains. (MP4 18435 kb)
Rights and permissions
About this article
Cite this article
Urnavicius, L., Lau, C., Elshenawy, M. et al. Cryo-EM shows how dynactin recruits two dyneins for faster movement. Nature 554, 202–206 (2018). https://doi.org/10.1038/nature25462
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25462
This article is cited by
-
The eukaryotic-like characteristics of small GTPase, roadblock and TRAPPC3 proteins from Asgard archaea
Communications Biology (2024)
-
Cargo specificity, regulation, and therapeutic potential of cytoplasmic dynein
Experimental & Molecular Medicine (2024)
-
Interaction of the Zika virus with the cytoplasmic dynein-1
Virology Journal (2023)
-
Conserved roles for the dynein intermediate chain and Ndel1 in assembly and activation of dynein
Nature Communications (2023)
-
TRAK adaptors regulate the recruitment and activation of dynein and kinesin in mitochondrial transport
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.