Abstract
The ageing suppressor α-klotho binds to the fibroblast growth factor receptor (FGFR). This commits FGFR to respond to FGF23, a key hormone in the regulation of mineral ion and vitamin D homeostasis. The role and mechanism of this co-receptor are unknown. Here we present the atomic structure of a 1:1:1 ternary complex that consists of the shed extracellular domain of α-klotho, the FGFR1c ligand-binding domain, and FGF23. In this complex, α-klotho simultaneously tethers FGFR1c by its D3 domain and FGF23 by its C-terminal tail, thus implementing FGF23–FGFR1c proximity and conferring stability. Dimerization of the stabilized ternary complexes and receptor activation remain dependent on the binding of heparan sulfate, a mandatory cofactor of paracrine FGF signalling. The structure of α-klotho is incompatible with its purported glycosidase activity. Thus, shed α-klotho functions as an on-demand non-enzymatic scaffold protein that promotes FGF23 signalling.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Shimada, T. et al. Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Invest. 113, 561–568 (2004)
Gattineni, J. et al. FGF23 decreases renal NaPi-2a and NaPi-2c expression and induces hypophosphatemia in vivo predominantly via FGF receptor 1. Am. J. Physiol. Renal Physiol. 297, F282–F291 (2009)
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010)
Schlessinger, J. et al. Crystal structure of a ternary FGF-FGFR-heparin complex reveals a dual role for heparin in FGFR binding and dimerization. Mol. Cell 6, 743–750 (2000)
Mohammadi, M., Olsen, S. K. & Ibrahimi, O. A. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 16, 107–137 (2005)
Goetz, R. & Mohammadi, M. Exploring mechanisms of FGF signalling through the lens of structural biology. Nat. Rev. Mol. Cell Biol. 14, 166–180 (2013)
Kuro-o, M. et al. Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51 (1997)
Henrissat, B. & Davies, G. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7, 637–644 (1997)
Goetz, R. et al. Klotho coreceptors inhibit signaling by paracrine fibroblast growth factor 8 subfamily ligands. Mol. Cell. Biol. 32, 1944–1954 (2012)
Goetz, R. et al. Isolated C-terminal tail of FGF23 alleviates hypophosphatemia by inhibiting FGF23–FGFR–Klotho complex formation. Proc. Natl Acad. Sci. USA 107, 407–412 (2010)
Urakawa, I. et al. Klotho converts canonical FGF receptor into a specific receptor for FGF23. Nature 444, 770–774 (2006)
Kurosu, H. et al. Regulation of fibroblast growth factor-23 signaling by klotho. J. Biol. Chem. 281, 6120–6123 (2006)
Li, S. A. et al. Immunohistochemical localization of Klotho protein in brain, kidney, and reproductive organs of mice. Cell Struct. Funct. 29, 91–99 (2004)
van Loon, E. P. et al. Shedding of klotho by ADAMs in the kidney. Am. J. Physiol. Renal Physiol. 309, F359–F368 (2015)
Lindberg, K. et al. The kidney is the principal organ mediating klotho effects. J. Am. Soc. Nephrol. 25, 2169–2175 (2014)
Chen, C. D., Podvin, S., Gillespie, E., Leeman, S. E. & Abraham, C. R. Insulin stimulates the cleavage and release of the extracellular domain of Klotho by ADAM10 and ADAM17. Proc. Natl Acad. Sci. USA 104, 19796–19801 (2007)
Imura, A. et al. Secreted Klotho protein in sera and CSF: implication for post-translational cleavage in release of Klotho protein from cell membrane. FEBS Lett. 565, 143–147 (2004)
Matsumura, Y. et al. Identification of the human klotho gene and its two transcripts encoding membrane and secreted klotho protein. Biochem. Biophys. Res. Commun. 242, 626–630 (1998)
Shiraki-Iida, T. et al. Structure of the mouse klotho gene and its two transcripts encoding membrane and secreted protein. FEBS Lett. 424, 6–10 (1998)
Kurosu, H. et al. Suppression of aging in mice by the hormone Klotho. Science 309, 1829–1833 (2005)
Hu, M. C., Shiizaki, K., Kuro-o, M. & Moe, O. W. Fibroblast growth factor 23 and Klotho: physiology and pathophysiology of an endocrine network of mineral metabolism. Annu. Rev. Physiol. 75, 503–533 (2013)
Chang, Q. et al. The β-glucuronidase klotho hydrolyzes and activates the TRPV5 channel. Science 310, 490–493 (2005)
Cha, S. K. et al. Removal of sialic acid involving Klotho causes cell-surface retention of TRPV5 channel via binding to galectin-1. Proc. Natl Acad. Sci. USA 105, 9805–9810 (2008)
Hu, M. C. et al. Klotho: a novel phosphaturic substance acting as an autocrine enzyme in the renal proximal tubule. FASEB J. 24, 3438–3450 (2010)
Imura, A. et al. α-Klotho as a regulator of calcium homeostasis. Science 316, 1615–1618 (2007)
Hayashi, Y. et al. Klotho-related protein is a novel cytosolic neutral beta-glycosylceramidase. J. Biol. Chem. 282, 30889–30900 (2007)
Andrukhova, O. et al. Klotho lacks an FGF23-independent role in mineral homeostasis. J. Bone Miner. Res. 32, 2049–2061 (2017)
Wu, X. et al. C-terminal tail of FGF19 determines its specificity toward Klotho co-receptors. J. Biol. Chem. 283, 33304–33309 (2008)
Ornitz, D. M. et al. Heparin is required for cell-free binding of basic fibroblast growth factor to a soluble receptor and for mitogenesis in whole cells. Mol. Cell. Biol. 12, 240–247 (1992)
Pellegrini, L., Burke, D. F., von Delft, F., Mulloy, B. & Blundell, T. L. Crystal structure of fibroblast growth factor receptor ectodomain bound to ligand and heparin. Nature 407, 1029–1034 (2000)
Harmer, N. J. et al. Towards a resolution of the stoichiometry of the fibroblast growth factor (FGF)–FGF receptor–heparin complex. J. Mol. Biol. 339, 821–834 (2004)
Ogawa, Y. et al. βKlotho is required for metabolic activity of fibroblast growth factor 21. Proc. Natl Acad. Sci. USA 104, 7432–7437 (2007)
Kurosu, H. et al. Tissue-specific expression of βKlotho and fibroblast growth factor (FGF) receptor isoforms determines metabolic activity of FGF19 and FGF21. J. Biol. Chem. 282, 26687–26695 (2007)
Holt, J. A. et al. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev. 17, 1581–1591 (2003)
Potthoff, M. J. et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc. Natl Acad. Sci. USA 106, 10853–10858 (2009)
Pitteloud, N. et al. Digenic mutations account for variable phenotypes in idiopathic hypogonadotropic hypogonadism. J. Clin. Invest. 117, 457–463 (2007)
White, K. E. et al. Autosomal dominant hypophosphataemic rickets is associated with mutations in FGF23. Nat. Genet. 26, 345–348 (2000)
Kabsch, W. Xds. Acta Crystallogr. D 66, 125–132 (2010)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 (2011)
Adams, P. D . et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 (2010)
Goetz, R. et al. Molecular insights into the klotho-dependent, endocrine mode of action of fibroblast growth factor 19 subfamily members. Mol. Cell. Biol. 27, 3417–3428 (2007)
Plotnikov, A. N., Schlessinger, J., Hubbard, S. R. & Mohammadi, M. Structural basis for FGF receptor dimerization and activation. Cell 98, 641–650 (1999)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Suzuki, M. et al. βKlotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol. Endocrinol. 22, 1006–1014 (2008)
Ornitz, D. M. et al. FGF binding and FGF receptor activation by synthetic heparan-derived di- and trisaccharides. Science 268, 432–436 (1995)
Liu, Y . et al. Regulation of receptor binding specificity of FGF9 by an autoinhibitory homodimerization. Structure 25, 1325–1336 (2017)
Belov, A. A. & Mohammadi, M. Molecular mechanisms of fibroblast growth factor signaling in physiology and pathology. Cold Spring Harb. Perspect. Biol. 5, a015958 (2013)
Beenken, A., Eliseenkova, A. V., Ibrahimi, O. A., Olsen, S. K. & Mohammadi, M. Plasticity in interactions of fibroblast growth factor 1 (FGF1) N terminus with FGF receptors underlies promiscuity of FGF1. J. Biol. Chem. 287, 3067–3078 (2012)
Olsen, S. K. et al. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev 20, 185–198 (2006)
Robinson, C. J., Harmer, N. J., Goodger, S. J., Blundell, T. L. & Gallagher, J. T. Cooperative dimerization of fibroblast growth factor 1 (FGF1) upon a single heparin saccharide may drive the formation of 2:2:1 FGF1.FGFR2c.heparin ternary complexes. J. Biol. Chem. 280, 42274–42282 (2005)
Goodger, S. J. et al. Evidence that heparin saccharides promote FGF2 mitogenesis through two distinct mechanisms. J. Biol. Chem. 283, 13001–13008 (2008)
Dunshee, D. R. et al. Fibroblast activation protein cleaves and inactivates fibroblast growth factor 21. J. Biol. Chem. 291, 5986–5996 (2016)
Acknowledgements
We thank N. J. Cowan for critically reading and editing the manuscript, and C.-S. Huang for help with diffraction data processing with XDS. This work was primarily supported by NIH grant R01 DE13686 (to M.M.). Support was also provided by National Key R&D Program of China (#2017YFA0506000 to X.L.). Funding for mouse studies was provided by R01 DK092461, P30 DK079328 (to O.W.M.), and R01 DK091392 (to M.C.H). Beamlines at the Northeastern Collaborative Access Team (NE-CAT) facility at the Advanced Photon Source of Argonne National Laboratory are primarily funded by NIH NIGMS and member institutions.
Author information
Authors and Affiliations
Contributions
G.C. purified and crystallized the ternary complex, analysed the crystal structure, generated SEC–MALS data (Figs 4a, 5a, b, f), cell-based data (Fig. 4), enzyme and thermostability assay data (Fig. 2c), and participated in the design of experiments and the writing/revising of the manuscript. Y.L. helped with data collection and analysis of the crystal structure, generated cell-based data (Fig. 5), and participated in manuscript revision. R.G. established expression and purification protocols for the ternary complex, performed ternary complex characterization, analysed mouse data, and participated in editing and revising the manuscript. L.F. generated expression constructs for FGF23, FGFR1cecto, α-klothoecto and their structure-based mutated forms, and helped with ternary complex purification. S.J. assisted with diffraction data collection and performed excitation/emission scanning of the FGF23–FGFR1cecto–α-klothoecto crystal (Extended Data Fig. 2c). M.-C.H. and O.W.M. generated the mouse data (Extended Data Figs 1c, d and 7a, b). G.L. and X.L. (mentors of G.C. and L.F.) participated in manuscript revision. M.M. developed and directed the project, solved, refined, analysed and interpreted the crystal structure of the ternary complex, and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
O.W.M. has done paid consultation for AbbVie, Allena, Amgen, and Tricida. He also sits on the board of Klotho Therapeutics. All of the other authors have no competing financial interests to declare.
Additional information
Reviewer Information Nature thanks M. Kuro-o and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
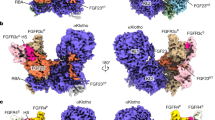
Extended Data Figure 1 α-Klothoecto functions as a co-receptor for FGF23.
a, Domain organization of membrane-bound α-klotho (α-klothoTM) and its soluble isoform α-klothoecto generated by an ectodomain shedding in the kidney16. KL1 and KL2 are tandem domains with homology to family 1 glycosidases8. b, Representative immunoblots of phosphorylated ERK (top) and total ERK (bottom; sample loading control) in total HEK293 cell lysates (n = 3 independent experiments). Top, lysates from untransfected HEK293 cells that were pre-treated with a fixed α-klothoecto concentration (10 nM) and then stimulated with increasing FGF23 concentrations, and lysates from HEK293-α-klothoTM cells treated with increasing concentrations of FGF23 alone. Bottom, lysates from HEK293-α-klothoTM cells that were pre-treated with increasing α-klothoecto concentrations and then stimulated with a fixed FGF23 concentration. c, Plasma phosphate, fractional excretion of phosphate, and phosphate excretion rate in wild-type mice before and after a single injection of α-klothoecto (0.1 mg kg−1 body weight) or isotonic saline alone (buffer). Circles denote mean values; error bars denote s.d. n = 10 mice per group. *P < 0.05, paired Student’s t test. d, Relative Egr1 mRNA levels in the kidney of wild-type mice after a single injection with α-klothoecto (0.1 mg kg−1 body weight) or isotonic saline alone (buffer). Data are mean and s.d. n = 3 mice per group. The same batch of α-klothoecto protein was used in the experiments shown in b–d.
Extended Data Figure 2 Topology of ternary complex is consistent with its orientation on the cell surface.
a, Cartoon representation of 1:1:1 FGF23–FGFR1cecto–α-klothoecto complex in four different orientations related by 90° rotation. α-Klotho domains are coloured cyan (KL1) and blue (KL2); the KL1–KL2 linker is in yellow. FGFR1c and FGF23 are in green and orange, respectively. The ternary complex resembles an oblique rectangular prism with an average dimension of 100 Å × 90 Å × 50 Å. The long axes of α-klothoecto and FGF23–FGFR1c complex in the ternary complex are each about 90 Å long, and parallel to one another such that the C termini of FGFR1cecto and α-klothoecto end up on the same side of the ternary complex, ready to insert into the cell membrane (grey bar). First, the N-acetyl glucosamine moiety (purple sticks) at six of the seven consensus N-linked α-klotho glycosylation sites could be built owing to sufficient electron density. Asn694 is the only glycosylation site that falls in the vicinity of a binding interface, namely α-klothoecto–FGF23. b, Close-up view of the KL1–KL2 interdomain interface. Zinc (orange sphere)-mediated contacts facilitate overall α-klothoecto conformation. Dashed yellow lines denote hydrogen bonds; grey surfaces denote hydrophobic contacts. c, Emission energy spectrum obtained from excitation/emission scan of the FGF23–FGFR1cecto–α-klothoecto crystal. Inset shows an expanded view of zinc fluorescence at 8,637 eV of emission energy.
Extended Data Figure 3 Structural basis for the weak FGFR-binding affinity of FGF23.
a, Open-book view of FGF23–FGFR1cecto complex interface. FGF23 (orange) and FGFR1cecto (green) are pulled apart and rotated by 90° around the vertical axis to expose the binding interface (blue). b, Ligand–receptor D3 and ligand–receptor D2–D3 linker interfaces of endocrine FGF23–FGFR1c and paracrine FGF9–FGFR1c47 structures. Grey transparent surfaces denote hydrophobic interactions; dashed yellow lines denote hydrogen bonds. Because FGF9 Arg62 is replaced with glycine in FGF23 (Gly38) and FGF9 Glu138 is replaced with histidine in FGF23 (His117), neither the side chain of Asp125 in FGF23 (Asn146 in FGF9), nor the side chain of invariant Arg250 in the FGFR1c D2–D3 linker can be tethered through intramolecular hydrogen bonds. Thus, these side chains possess greater freedom of motion in the FGF23–FGFR1c complex, and as a result, hydrogen bonding between FGF23 and FGFR1c D2–D3 linker entails greater entropic cost, which generates less binding affinity. Substitution of Phe140 and Pro189 in FGF9 with hydrophilic Thr119 and Ser159 in FGF23 further diminishes the ability of FGF23 to gain binding affinity from hydrogen bonding with FGFR1c D2–D3 linker. A lack of contacts between FGF23 N terminus and FGFR1c D3 cleft, which forms between alternatively spliced βC′-βE and βB′-βC loops48, probably further exacerbates the weak FGFR-binding affinity of FGF23. c, Ligand–receptor D2 interface in endocrine FGF23–FGFR1c and paracrine FGF9–FGFR1c47 structures. Grey transparent surfaces denote hydrophobic interactions; dashed yellow lines denote hydrogen bonds. Many contacts at this interface are conserved between paracrine FGF molecules and FGF23, and hence FGF23 gains much of its FGFR-binding affinity through these contacts. Three hydrogen bonds involving Asn49, Ser50 and His66 of FGF23 are unique to the FGF23–FGFR1c complex.
Extended Data Figure 4 Structural basis for FGFR isoform specificity of a-klotho and FGF23.
a, Structure-based sequence alignment of a segment of FGFR D3. The alternatively spliced regions of all seven FGFRs are boxed with a purple rectangle. β-strand locations above the alignment are coloured green (constant region) and purple (alternatively spliced region). A leucine (boxed) of hydrophobic groove residues (light purple) in the alternatively spliced region is conserved only among ‘c’ isoforms of FGFR1–FGFR3 and FGFR4, which explains α-klotho binding selectivity for these receptors. b, Interface between FGF23 and the βF–βG loop of FGFR1c D3 in the FGF23–FGFR1c structure of the ternary complex. Backbone atoms of His117 and Gly81 in FGF23 make specific hydrogen bonds with the Ser346 side-chain and Asn345 backbone atoms of the βF–βG loop. The serine residue corresponding to Ser346 in FGFR1c (yellow) is conserved only among ‘c’ isoforms of FGFR1–FGFR3 and FGFR4 (see a). c, Representative immunoblots of phosphorylated ERK (top) and total ERK (bottom; sample loading control) in total BaF3 cell lysates (n = 3 independent experiments). d, Cartoon representations of four paracrine FGF–FGFR complex structures4,47,49,50. Solid black oval denotes the hydrophobic D3 groove. Dashed black circle denotes the second binding pocket (SBP) for α-klotho in D3. Although the hydrophobic groove is engaged by FGF8 (see also e), the SBP is not used in any of the current paracrine FGF–FGFR structures. In most paracrine FGF–FGFR structures, the βC-βC′ loop is disordered (dashed red lines) because it does not participate in FGF binding. Evidently, SBP and βC-βC′ loop in D3 have evolved to mediate α-klotho binding to FGFR. e, α-Klotho and FGF8b both bind to the hydrophobic groove in FGFR1c D3. FGF8b (brown) from the FGF8b–FGFR2c structure50 was superimposed onto FGF23 in the FGF23–FGFR1cecto–α-klothoecto complex. The αN helix of FGF8b occupies the same binding pocket in FGFR1c D3 as the distal tip of the α-klotho RBA.
Extended Data Figure 5 α-Klotho is the first non-enzymatic scaffold among TIM barrel proteins.
a, Structure-based sequence alignment of TIM barrels of α-klotho KL1 and KL2 domains and KLrP. Most glycoside hydrolases (GH), a functionally diverse group of enzymes that cleave glycosidic bonds of complex carbohydrates on glycoproteins8, adopt a TIM barrel fold. Locations and lengths of TIM barrel β-strands and α-helices are indicated above the alignment. Among GH family 1 members of the klotho subfamily, only KLrP has a verified glycosylceramidase activity26, and Glu165 and Glu373 are its catalytically essential glutamic acids. KLrP residues coloured cyan participate in substrate recognition/hydrolysis. α-Klotho residues coloured red bind FGF23, and α-klotho residues of the KL2 β1α1 loop (purple box) coloured purple interact with the FGFR1c D3 domain. b, Superimposition of KL1 Cα trace (grey/cyan) onto that of KLrP (grey/yellow). Superimposition root mean square deviation (r.m.s.d.) value is 1.08 Å. Structurally most divergent regions between KL1 and KLrP are in cartoon representation. Glucose moiety and aliphatic chains of glucosylceramide (KLrP substrate) are in sticks with carbon in black (glucose) or green/cyan/pink (aliphatic chains). Catalytically essential Glu165 in KLrP is replaced by an asparagine in KL1. Hydrophobic residues from KL1 β6α6 loop occupy the pocket that accommodates the aliphatic chains of glucosylceramide in KLrP. The KL1 N terminus supports KL1–KL2 cleft formation (Extended Data Fig. 2b) and KL1 β6α6 loop conformation contributes to a key portion of the binding pocket in this cleft for the FGF23 C-terminal tail (Fig. 3c). c, d, Superimposition of KL2 Cα trace (grey/blue) onto that of KLrP (grey/yellow). Superimposition r.m.s.d. value is 1.37 Å. Structurally divergent β1α1 (c), β6α6 and β8α8 (d) loops of KL2 and KLrP are rendered in cartoon. β1α1 loop in KL2 is disengaged from the central TIM barrel and stretches away from it by as much as 35 Å. Catalytically essential Glu373 in KLrP is replaced by a serine in KL2. KLrP residues from β6α6 and β8α8 loops bind glucosylceramide (KLrP substrate); for example, Trp345 in the β6α6 loop and Glu424 and Trp425 in the β8α8 loop. Sequence divergence (a) and altered loop conformations are incompatible with glucosylceramide coordination by KL2. β1α1, β6α6 and β8α8 loops lie at the rim of the catalytic mouth in the TIM barrel (see Fig. 2b). Divergent conformations of these three loops in KL2 result in notable widening of the central barrel cavity in KL2, which merges with the KL1–KL2 cleft to form an expansive basin that accommodates the distal portion of the FGF23 C-terminal tail.
Extended Data Figure 6 α-Klotho interaction with rigid core of FGF23 and a second binding pocket next to the hydrophobic groove in FGFR1c D3.
a, A partial view of the ternary complex. α-Klothoecto (cyan/blue solid surface, RBA of KL2 in blue cartoon), FGF23 (orange transparent surface and cartoon), FGFR1c (constant region: solid green surface; alternatively spliced region: solid purple surface). Dashed black circle denotes the perimeter of the interface between proximal end of α-klotho RBA and a second binding pocket (SBP) in FGFR1c D3 next to the hydrophobic groove. Solid black box denotes the perimeter of α-klotho−FGF23core interface. b, Close-up view of the interface between proximal end of RBA and SBP in D3. The disulfide bridge between Cys572 (N-terminal end of RBA) and Cys621 (α2 helix) at the base of the RBA probably imparts some degree of conformational rigidity to the proximal RBA portion, whereas the conformation of the distal RBA tip is dictated by contacts with FGFR1c D3. c, Close-up view of the α-klotho−FGF23core interface detailing hydrogen bonding (top) and hydrophobic contacts (bottom). Grey transparent surfaces denote hydrophobic interactions; dashed yellow lines denote hydrogen bonding contacts.
Extended Data Figure 7 Deletion of RBA of α-klothoecto generates an FGF23 ligand trap.
a, Plasma phosphate and fractional excretion of phosphate in wild-type mice before and after a single injection of α-klothoecto (0.1 mg kg−1 body weight), mutant α-klothoecto/ΔRBA (0.1 mg kg−1 body weight), or isotonic saline alone (buffer). Circles denote mean values; error bars denote s.d. n = 6 mice per group. Significance values were determined by a paired Student’s t test. b, Relative Egr1 mRNA levels in the kidney of wild-type mice injected once with α-klothoecto (0.1 mg kg−1 body weight; n = 3), mutant α-klothoecto/ΔRBA (0.1 mg kg−1 body weight; n = 4), or isotonic saline alone (buffer; n = 3). Data are mean and s.d. c, Representative elution profiles of FGF23–α-klothoecto and FGF23–α-klothoecto/ΔRBA mixtures from a size-exclusion column and representative Coomassie blue-stained SDS-polyacrylamide gels of eluted protein peak fractions. d, Thermal shift assay of α-klothoecto and the α-klothoecto/ΔRBA mutant in the presence and absence of FGF23 C-terminal tail peptide (FGF23C-tail) (n = 3 independent experiments). Increased melting temperatures in the presence of the FGF23C-tail indicate interaction of both α-klothoecto proteins with the peptide. Higher melting temperature of α-klothoecto/ΔRBA mutant relative to wild-type α-klothoecto indicates greater stability of the mutant protein. e, Representative immunoblots of phosphorylated ERK (top) and total ERK (bottom; sample loading control) in total lysates from HEK293-α-klothoTM cells co-stimulated with a fixed FGF23 concentration and increasing α-klothoecto/ΔRBA concentrations (n = 3 independent experiments). The α-klothoecto/ΔRBA mutant inhibits FGF23-induced ERK phosphorylation owing to sequestering FGF23 into inactive FGF23–α-klothoecto/ΔRBA binary complexes. This also explains why α-klothoecto/ΔRBA injection into mice causes an increase in plasma phosphate (a) concomitant with renal Egr1 gene repression (b).
Extended Data Figure 8 FGF23–FGFR1cecto–α-klothoecto–HS quaternary dimer models.
a, A 2:2:2:1 FGF23–FGFR1cecto–α-klothoecto–HS quaternary dimer in two orientations related by a 90° rotation around the horizontal axis. The dimer was constructed by superimposing FGF23 from two copies of 1:1:1 FGF23–FGFR1cecto–α-klothoecto complex onto the two FGF1 molecules in the 2:2:1 FGF1–FGFR2c–HS dimer30,31,51,52. The dimer is held together solely by HS, which bridges two FGF23 molecules in trans. Boxed pink surface denotes the location of Ala171, Ile203 and Val221 of FGFR1c, the mutation of which impairs the ability of HS to induce 2:2:2:2 quaternary dimer formation (Fig. 5f). Boxed grey region denotes the location of Met149, Asn150 and Pro151 of FGF23, the mutation of which diminishes HS-induced quaternary dimerization (Fig. 5e, f). None of these residues has any role in 2:2:2:1 quaternary dimer formation, and hence, contrary to experimental evidence (Fig. 5), mutation of these residues should not affect HS-induced FGF23–FGFR1cecto–α-klothoecto dimerization. b, A 2:2:2:2 FGF23–FGFR1cecto–α-klothoecto–HS quaternary dimer in two orientations related by a 90° rotation around the horizontal axis. See also Fig. 5g. The dimer was constructed by superimposing FGF23 from two copies of 1:1:1 FGF23–FGFR1cecto–α-klothoecto complex onto the two FGF2 molecules in the 2:2:2 FGF2–FGFR1c–HS dimer4. Insets show close-up views of the secondary FGF–FGFR (top) and direct FGFR–FGFR (bottom) interfaces. Grey/pink transparent surfaces denote hydrophobic interactions. Mutation of Ala171, Ile203 and Val221 (pink) impairs the ability of HS to dimerize the FGF23–FGFR1cecto–α-klothoecto ternary complex (Fig. 5f).
Extended Data Figure 9 The FGF19 and FGF21 co-receptor β-klotho is a non-enzymatic scaffold protein analogous to α-klotho.
Structure-based sequence alignment of α-klotho and β-klotho. The locations of the eight alternating β-strands and α-helices of the TIM fold are indicated above the alignment. Cyan, blue and yellow bars below the alignment mark the domain boundaries of KL1, KL2 and the KL1–KL2 linker. Asterisks denote sequence identity and dots denote sequence similarity. Scissor symbols mark the four proposed sites of α-klotho cleavage by ADAM proteases/secretases. Cleavage 1, which coincides with the end of the rigid core of KL2, results in shedding of the entire α-klotho ectodomain from the cell membrane. Although this cleavage product is a functional co-receptor, the α-klotho fragments generated by cleavages 2, 3 and 4 would be devoid of co-receptor activity. Black triangle denotes the site where alternative splicing replaces the C-terminal KL2 sequence with a 15-residue-long unrelated sequence. Glycan chain symbols denote seven predicted N-linked glycosylation sites. Zn2+-chelating residues of α-klotho are green, FGFR1c-binding residues are light purple, and FGF23-binding residues are red. Light purple box denotes β1α1 loop sequence in KL2 termed RBA. β-Klotho RBA is about as long as α-klotho RBA, and key FGFR-binding residues are conserved between these two RBAs, which is consistent with the similar FGFR-binding specificity of α-klotho and β-klotho9,11,12. But α-klotho residues in the binding pockets for the FGF23 C-terminal tail are not conserved in β-klotho, conforming to major sequence differences between the C-terminal tails of FGF23, FGF19 and FGF21 (Extended Data Fig. 10a).
Extended Data Figure 10 β-Klotho-dependent FGFR activation by FGF19 and FGF21 is mechanistically similar to α-klotho-dependent FGFR activation by FGF23.
a, Structure-based sequence alignment of endocrine FGF proteins. β-strands and the αC helix comprising the atypical β-trefoil core of FGF23 are indicated above the alignment. Asterisks and dots below the alignment denote sequence identity and similarity, respectively. Scissor symbols mark inactivating proteolytic cleavage sites in FGF23 and FGF2153. RXXR cleavage motif in FGF23 is in green bold letters. FGFR1c-binding residues of FGF23 are coloured blue, α-klotho-binding residues are coloured red. Vertical blue arrow marks the C-terminal boundary of the FGF23 variant used to solve the FGF23–FGFR1cecto–α-klothoecto complex structure. Five residues at the distal C-terminal region of FGF19 or FGF21 (black and grey) mediate binding of FGF19 or FGF21 to β-klotho. These residues completely diverge from the α-klotho-binding residues in the FGF23 C-terminal tail. α-Klotho-binding residues in the FGF23 core also are not conserved in FGF19 and FGF21. b, Representative immunoblots of phosphorylated ERK (top) and total ERK (bottom; sample loading control) in total lysates from HEK293 cells expressing wild-type or mutant β-klothoTM (n = 3 independent experiments). Similar to α-klothoΔRBA, β-klothoΔRBA failed to support FGF21-induced FGFR activation, and β-klotho (L394P) and β-klotho (M435Y) mutants also had greatly diminished ability to promote FGF21 signalling. Thus, β-klotho tethers FGFR1c and FGF21 to itself in a manner similar to that identified for α-klotho to enable FGF21 signalling. c, Representative immunoblots of phosphorylated ERK (top) and total ERK (bottom; sample loading control) in total lysates from BaF3 cells expressing FGFR1c and β-klothoTM (n = 3 independent experiments). Like α-klotho, β-klotho also requires heparin to support FGF21-mediated FGFR1c activation.
Supplementary information
Supplementary Information
This file contains Supplementary Figure 1 and Supplementary Tables 1-2. (PDF 1058 kb)
Rights and permissions
About this article
Cite this article
Chen, G., Liu, Y., Goetz, R. et al. α-Klotho is a non-enzymatic molecular scaffold for FGF23 hormone signalling. Nature 553, 461–466 (2018). https://doi.org/10.1038/nature25451
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature25451
This article is cited by
-
Relationships of serum FGF23 and α-klotho with atherosclerosis in patients with type 2 diabetes mellitus
Cardiovascular Diabetology (2024)
-
FGF23 and klotho at the intersection of kidney and cardiovascular disease
Nature Reviews Cardiology (2024)
-
Revealing the tumor suppressive sequence within KL1 domain of the hormone Klotho
Oncogene (2024)
-
Overexpression of α-Klotho isoforms promotes distinct Effects on BDNF-Induced Alterations in Dendritic Morphology
Molecular Neurobiology (2024)
-
Serum klotho levels and mortality patterns in frail individuals: unraveling the u-shaped association
Aging Clinical and Experimental Research (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.